Figure 4.
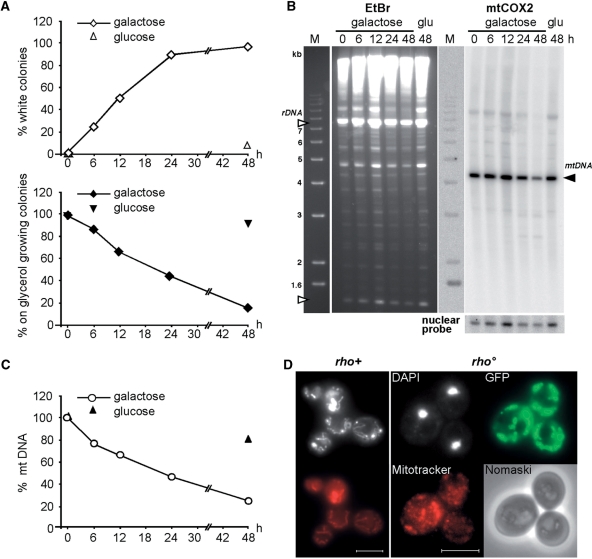
Mitochondrially targeted mt125Top1-103 leads to mtDNA loss and the formation of rho° petite. (A) Time course of mt125Top1-103 dependent petite formation. Cells were grown in galactose-containing liquid medium (expression). At the indicated times cells were retrieved, grown on glucose containing plates (no expression) and replica-plated onto glycerol. Petite cells were determined by color (white; open diamonds) and their inability to grow on glycerol (black diamonds). Average values of two independent experiments are shown. (B) CHEF analysis of the mtDNA copy number. In-plug isolated, total cellular DNA was ApaI digested, gel electrophoresed and analyzed by Southern blot. Shown are the agarose gel before Southern blotting (rDNA fragments, open triangle) and the signals obtained using a specific probe against mitochondrial (right top, black triangle) and nuclear DNA (right bottom) after Southern blotting. (C) Determination of the relative mtDNA copy number. The mtCOX2 hybridization signal obtained at time point 0 was normalized to the nuclear probe and set as 100%. Average values of two independent experiments are shown. (D) Wide-field fluorescence microscopy analysis of petite cells after 48 h of mt125Top1-103 expression. DAPI (white) and Mitotracker (red) staining of rho+ (w/o induction) and rho° (48 h of induction) is shown. White bar represents 5 µm.