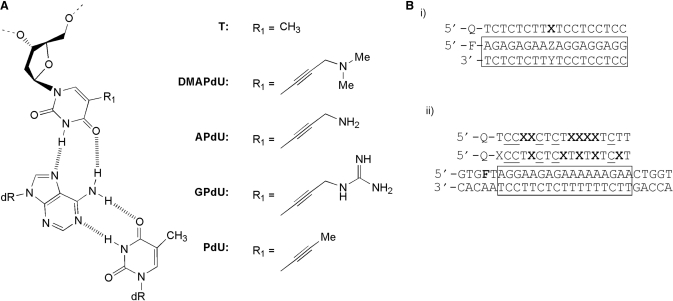
Figure 1.
(A) Chemical structures of the X.AT triplets, where X is either T or the 5-position deoxyuridine analogues examined in this study (B) (i) Sequences of the oligonucleotides used in single substitution fluorescence melting studies. The third strand is labelled with dabcyl (Q) at the 5′-end, whilst the purine strand of the duplex (boxed) is labelled with fluorescein (F) at the 5′-end. Each analogue was examined in turn at position X against duplex targets containing each base pair at position ZY. (ii) Sequence of the oligonucleotides used in multiple substitution fluorescence melting and DNase I footprinting studies. In melting experiments the purine strand of the duplex is labelled with fluorescein attached to T adjacent to the 5′-end of the oligopurine tract (F = FAM-dT). In footprinting experiment the TFO target (F = T) is contained within the 110 base pair tyrT(43–59) DNA fragment which was labelled at the 3′-end of the EcoRI site with [α-32P]dATP. In both cases the third strand was labelled with dabcyl (Q) at the 5′-end and contained methyl C (C) in place of C. Each analogue was examined in turn as six substitutions at position X in each TFO.