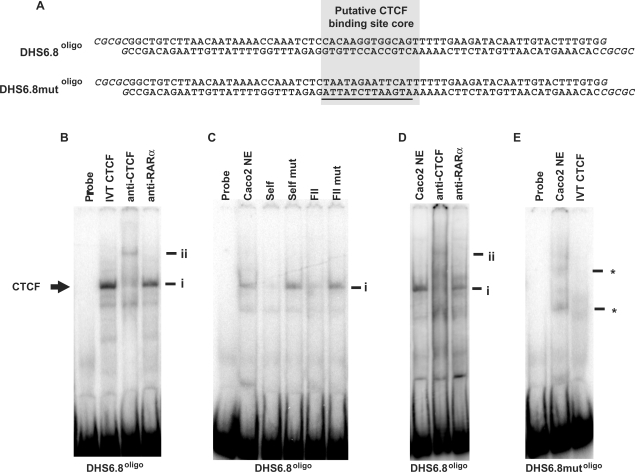
Figure 2.
In vitro binding of CTCF at the +6.8 kb DHS region. (A) The DHS6.8oligo and DHS6.8mutoligo probes. The putative CTCF-binding site core is highlighted. Oligonucleotides were designed to form BssHII sticky ends when annealed (shown in italics), facilitating cloning into the AscI site of pNI. Mutated bases in DHS6.8mutoligo are underlined. (B) IVT CTCF; (C, D), Caco2 nuclear extracts. (B) EMSA using DHS6.8oligo. Supershift was performed with an anti-CTCF antibody and anti-RAR β was used as isotype control. (C) EMSA with DHS6.8oligo. Competition reactions were performed with 100× excess of unlabelled DHS6.8oligo (self), DHS6.8mutoligo (self mut), FII (known CTCF-binding site from chicken β-globin locus) and mutant (FIImut). (D) EMSA with DHS6.8oligo. Supershift reactions were performed as in (B). (E) EMSA with DHS6.8mutoligo. Complex marked i represents CTCF in complex with DHS6.8oligo and ii the antibody-supershifted CTCF complex. Undefined interactions formed between 6.8mutoligo and Caco2 nuclear extract are marked by *.