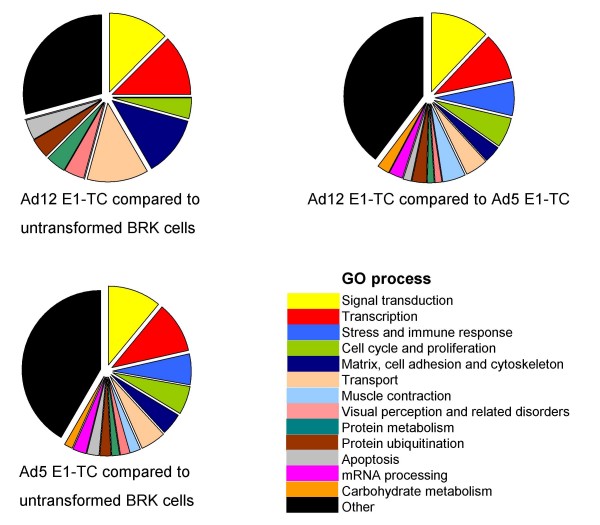
Figure 2.
Significantly dysregulated transcripts can be further characterised by gene ontology. Pie charts showing gene ontology analysis of the transcripts differentially expressed as a result of Ad transformation: Ad12 E1-TC compared to untransformed BRK cell line (neoplastic transformation); Ad12 E1-TC compared to Ad5 E1-TC (oncogenic transformation compared to non-oncogenic transformation) and Ad5 E1-TC compared to untransformed BRK cell line (non-oncogenic transformation). It was possible to classify the majority of genes in to categories defined by GO biological processes with reference to the DAVID, KEGG and GenMAPP databases (see Materials and Methods). The largest category is that containing gene products that play roles in signal transduction and transcription.