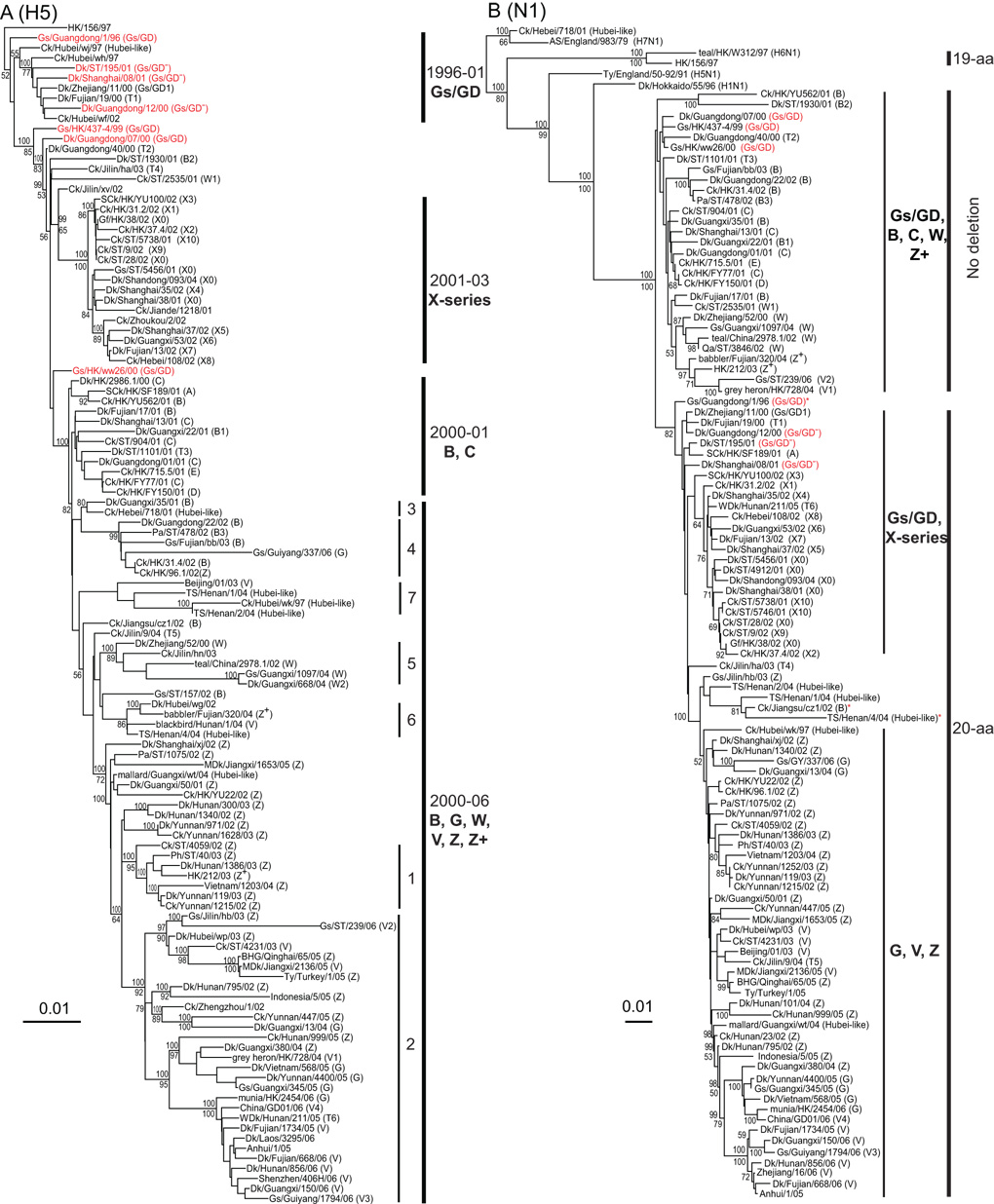
Fig. 1.
Phylogenetic relationships of the HA (A) and NA (B) genes of representative influenza A viruses. Analyses were based on nucleotides 1–1,696 of the HA gene and nucleotides 1–1,355 of the NA gene. The HA and NA gene trees were rooted to duck/Hokkaido/51/96 and chicken/Scotland/59, respectively. The numbers above and below the branch nodes indicate Bayesian posterior probabilities of ≥95 and neighbor-joining bootstrap values of ≥50%, respectively. H5N1 prototype virus names are in red. Virus genotypes are given in parenthesis following the virus names. Major genotypes are indicated with bold text. Numbers labeled on the HA tree indicate WHO H5N1 clade designations (http://www.who.int/csr/disease/avian_influenza/guidelines/nomenclature/en). In the NA tree, 19-aa and 20-aa indicate the presence of amino acid deletions in the stalk region. *Indicates viruses without the 20-aa NA deletion. Scale bar, 0.01 nucleotide substitutions per site. Abbreviations: AS, African starling; BHG, bar-headed goose; Ck, chicken; Dk, duck; Gf, Guinea fowl; Gs, goose; HK, Hong Kong; MDk, migratory duck; Pa, partridge; Ph, pheasant; SCk, silky chicken; ST, Shantou; TS, tree sparrow; Ty, Turkey; WDk, wild duck.