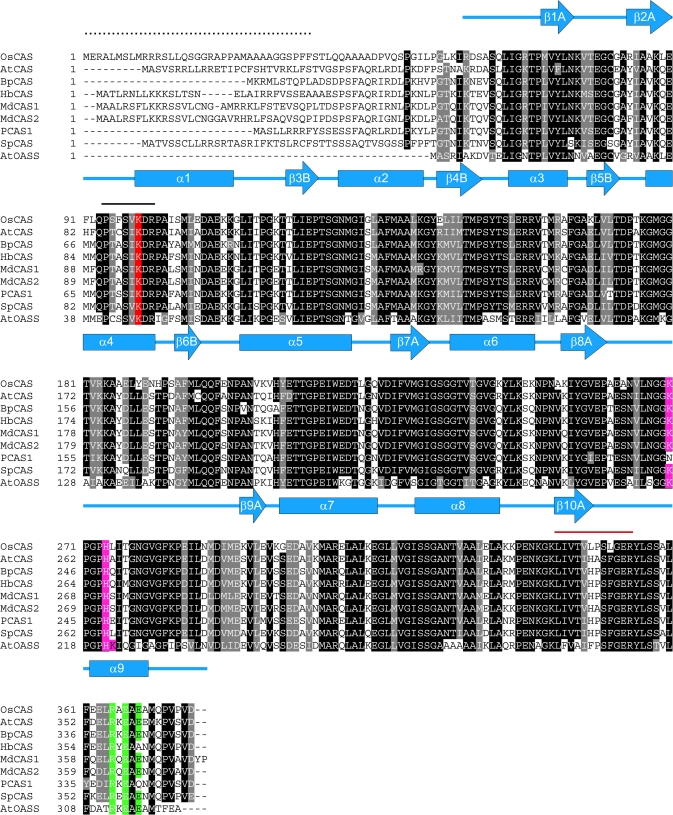
Fig. 1.
Alignment of sequences of OsCAS and other CAS from plant species with AtOASS from Arabidopsis. Comparison of the derived amino acid sequence of OsCAS was made with those of the Arabidopsis AtCAS (Hatzfeld et al., 2000), spinach SpCAS (Hatzfeld et al., 2000), potato PCAS1 (Maruyama et al., 2001), birch BpCAS (GeneBank accession number AY154650), latex (Hevea brasilienis) HbCAS (GeneBank accession number AAP41852), Fuji apple MdCAS1, MdCAS2 (Han et al., 2007), and Arabidopsis AtOASS (EMBL number CAA56593). Amino acid residues that are conserved in at least eight of the nine sequences are shown in black. Secondary structures of AtOASS are represented in α-helices (blue rectangles) and β-stands (blue arrows). The solid line in black shows the conserved pyridoxal 5′ phosphate binding site while the red line covers a peptide from the partially purified rice CAS fraction sequenced by MS/MS. The mitochondrial target sequence from OsCAS which was determined by N-terminal Edman degradation on a mature OsCAS protein purified from a heterologous expression of Arabidopsis plant is indicated by dots. Amino acid residues which were chosen in this mutagenesis study are highlighted: Lysine in the PLP-binding domain is shaded in red while conserved amino acid residues for SAT interaction and dimerization are shaded in purple and green, respectively.