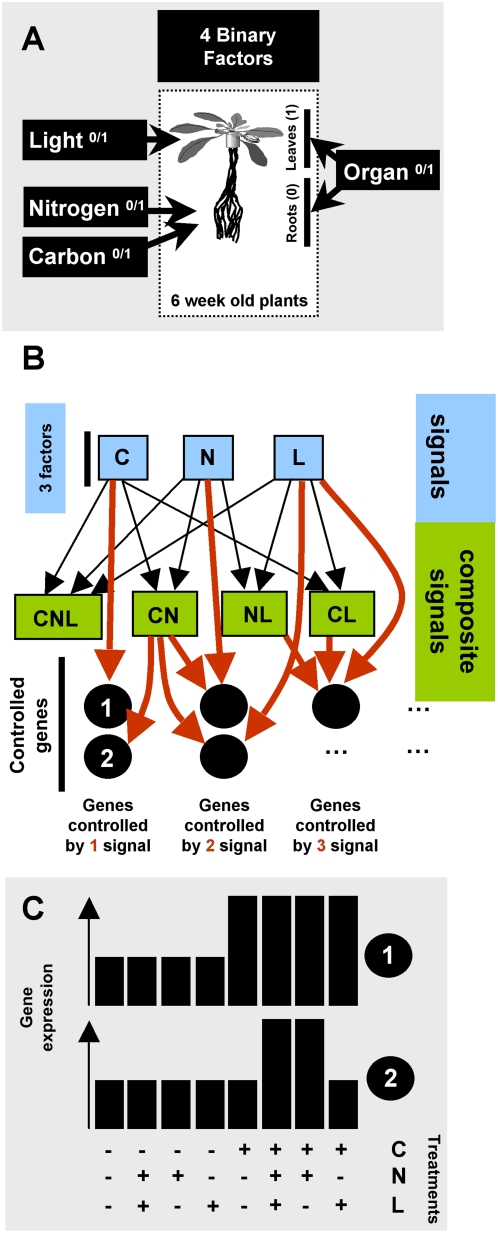
Figure 1. Scheme of experimental design and working model of gene control by multiple signals at the organ-specific level.
A) 6-week-old plants were treated for 8 h with all combinations of three (C, L, N) binary (0/1) factors. Leaves and roots were analyzed separately for a total of 16 experimental conditions. Treatments were as follows: N, 5 mM NO3 −; C, 30 mM sucrose; L, 60 µmol.m−2.s−1. RNAs were extracted from roots and shoots separately and hybridized to ATH1 Affymetrix chips. Microarray data analysis was performed as described in Experimental Procedures. B) Scheme presenting the concept used to decipher signal interactions in the control of gene expression. We propose that perceived signals can be produced from a factor (C, N, L represented as blue squares) or combination of factors (green squares). These combination of factors build what we name “composite signals”. These signals or composite signals can then affect the expression of a particular gene. The expression of a gene (e.g. black circles labeled 1 and 2) can be affected by (red arrow) one signal (e.g., C alone for number 1) or a composite signal (e.g., C and N for number 2). C) Idealized gene expression patterns produced by the signal effects shown in (B) for the genes 1 and 2.