Abstract
Biliary epithelial cells (cholangiocytes) respond to proinflammatory cytokines such as IFN-γ and actively participate in the regulation of biliary inflammatory response in the liver. B7-H1 (also known as CD274 or PD-L1) is a member of the B7 costimu-latory molecules and plays a critical immunoregulatory role in cell-mediated immune responses. In this study, we show that resting human cholangiocytes in culture express B7-H1 mRNA, but not B7-H1 protein. IFN-γ induces B7-H1 protein expression and alters the microRNA (miRNA) expression profile in cholangiocytes. Of those IFN-γ-down-regulated miRNAs, we identified microRNA-513 (miR-513) with complementarity to the 3′-untranslated region of B7-H1 mRNA. Targeting of the B7-H1 3′-untranslated region by miR-513 results in translational repression. Transfection of cholangiocytes with an antisense oligonucleotide to miR-513 induces B7-H1 protein expression. Additionally, transfection of miR-513 precursor decreases IFN-γ-induced B7-H1 protein expression and consequently influences B7-H1-associated apoptotic cell death in cocultured Jurkat cells. Thus, miR-513 regulates B7-H1 translation and is involved in IFN-γ-induced B7-H1 expression in human cholangiocytes, suggesting a role for miRNA-mediated gene silencing in the regulation of cholangiocyte response to IFN-γ.
Biliary epithelial cells (cholangiocytes) are the primary target cells in an important group of genetic and acquired hepatobiliary disorders collectively called “cholangiopa-thies” (1). Despite their heterogeneity, cholangiopathies share a number of basic pathogenetic mechanisms and the biliary inflammatory reaction is central to most manifestations of cholangiopathies (1-3). Considerable clinical data suggest that proinflammatory cytokines such as IFN-γ and TNF-α are key factors in driving persistent inflammation and cellular injury in patients with cholangiopathies (1-3). Indeed, cholangiocytes express receptors for IFN-γ and TNF-α (1). Proinflammatory cytokines mediate many cellular events in cholangiocytes including proliferation, apoptosis, cytotoxicity, cholestasis, and fibrosis (1).
B7-H1 (also known as CD274 or programmed death receptor-1 (PD-1) ligand-1) is a key member of the B7 costimulating molecules with important regulatory functions in cell-mediated immune responses (4). Although expression of B7-H1 mRNA is common in many cells, B7-H1 protein is usually undetectable, suggesting posttranscriptional suppression (4, 5). Proinflammatory cytokines such as IFN-γ and TNF-α are potent activators for inducing B7-H1 protein expression in T cells, B cells, endothelial cells, and epithelial cells (6). Cellular expression of B7-H1 is closely related to the clinical progress of a variety of important diseases such as HIV disease progression, autoimmune encephalomyelitis, and renal cell carcinoma (7-9). B7-H1-deficient mice show spontaneous accumulation of CD8+ T cells in the liver, development of autoimmune syndromes, and overreactions to microbial infections (6, 10, 11), suggesting a critical role for B7-H1 in the regulation of inflammatory responses in the liver. Whereas attention has been focused on its expression in lymphocytes, Kupffer cells, and sinusoidal endothelial cells in the liver (11, 12), more recent studies reveal a cholangiocyte-associated B7-H1 expression in patients with inflammatory hepatobiliary disorders (13).
MicroRNAs (miRNAs)4 are single-stranded regulatory RNA molecules of ∼21-23 nt (14, 15). miRNAs regulate gene expression based on their complementarity to the 3′-untranslated region (UTR) of target mRNAs resulting in mRNA cleavage and/or translational suppression (14, 16). It has been predicted that miRNAs control 20-30% of human genes (14). Over 700 miRNAs have been identified in human cells, including epithelial cells. Whereas most of the recent work involving miRNAs has been done in cancer- and development-related studies, miRNAs might be important players in the regulation of host immune response (16). Because miRNAs appear to provide quantitative regulation of genes rather than on-off decisions, they can be seen as a fine tuning for the cellular responses to external influences (17). Indeed, miRNAs have been implicated in the regulation of TLR signaling, viral immune escape, and antiviral defense (16, 17). Induction of miR-155 during the macrophage inflammatory response suggests its potential involvement in the regulation of inflammation (16, 17). We recently showed that a cellular miRNA, let-7i, is involved in translational regulation of TLR4 in human cholangiocytes (18). Nevertheless, whether miRNAs are involved in the posttranscriptional regulation of B7-H1 expression remains unclear.
In work described here, we showed that a cellular miRNA, microRNA-513 (miR-513), is expressed in human cholangiocytes and is down-regulated upon IFN-γ exposure. miR-513 is capable of targeting a predicted site in the B7-H1 3′-UTR, resulting in translational repression. Transfection of an antisense to miR-513 induces B7-H1 protein expression. miR-513 precursor transfection can reduce IFN-γ-stimulated B7-H1 expression and consequently influence B7-H1-associated apoptosis in cocultured T cells. Thus, a novel miR-513-mediated regulation of B7-H1 expression has been identified in cholangiocytes that may be associated with cholangiocyte inflammatory response in the liver and be relevant to the regulation of B7-H1 expression in general.
Materials and Methods
Cells
H69 cells are SV40-transformed immortalized human cholangiocytes originally derived from normal liver harvested for transplant and have been extensively characterized (19). Human intrahepatic biliary epithelial cells (HIBEpiC) are nonimmortalized, isolated human cholangiocytes commercial available from ScienCell Research Laboratories. HIBEpiC were grown on poly-l-lysine-coated dishes and cultured with the medium supplied from ScienCell Research Laboratories according to the manufacturer’s instructions. Jurkat cells were purchased from the American Type Culture Collection.
Western blotting
Whole cell lysates were obtained from HIBEpiC and H69 cells with MPER mammalian protein extraction reagent (Pierce) containing several protease inhibitors (1 mM PMSF, 10 μg/ml leupeptin, and 2 μg/ml pepstatin). Cell lysates were then loaded in SDS-PAGE gel to separate proteins and transferred to nitrocellulose membrane. Abs to B7-H1 (clone 5H1-A3, a gift from Dr. H. Dong at Mayo Clinic, Rochester, MN, and clone MIH1 from eBioscience), STAT1 (Cell Signaling), and actin (Sigma-Aldrich) were used. Densitometric levels of Western blot signals were quantified and expressed as their ratio to actin.
Small interfering RNA (siRNA) to STAT1
A siRNA specific to STAT1 and a nonspecific siRNA (i.e., scrambled siRNA as the control) were purchased from Cell Signaling. Cells were transfected with the siRNAs according to the manufacturer’s instruction and knockdown of STAT1 was confirmed by Western blotting.
miRCURY LNA array analysis of miRNAs
The Exiqon miRCURY LNA microRNA arrays and the service to process the samples were used. Briefly, H69 cells were grown to confluence and exposed to IFN-γ (10 ng/ml) for 8 h. Total RNAs were prepared with the mirVana miRNA isolation kit according to the manufacturer’s instruction (Ambion). The quality of isolated RNAs was verified by an Agilent 2100 Bioanalyzer profile, and a mixture of equal amounts of total RNAs from the control and IFN-γ-stimulated cells was used as the reference pool. A total of 2 μg of RNA from each sample was then labeled with an Hy5 fluorescent label and the reference pool was labeled with Hy3 using the miRCURY LNA Array labeling kit (Exiqon). The labeled samples and reference pool were then mixed pairwise and hybridized to the miRCURY LNA array containing capture probes targeting all human miRNAs listed in the miRBASE version 10.0 (Exiqon). After hybridization, the slides were scanned and quantified signals were normalized by Exiqon using the global LOWESS (locally weighted scatterplot smoothing) regression algorithm. Normalized Hy5/Hy3 ratios were used for further analysis as previously reported (20, 21).
PCR assay
For conventional RT-PCR analysis of mRNA expression, total RNA was isolated from cells with TRI Reagent (Ambion). Total RNA (1 μg) was reverse transcribed to cDNA by using a Moloney murine leukemia virus reverse transcriptase kit (Invitrogen). The primers used for PCR analysis were as follows: 5′-AAACAATTAGACCTGGCTG-3′ (forward) and 5′-TCTTACCACTCAGGACTTG-3′ (reverse) for human B7-H1; 5′-TGTGACGAGATTCAGTGCCAG-3′ (forward) and 5′-TCGGTATGCATGCCTGGAAC-3′ (reverse) for IFN-γR1; 5′-GAAGATTCGCCTGTACAACGC-3′ (forward) and 5′-TTCCAAAGCAGTTGTGCCTG-3′ (reverse) for IFN-γR2; 5′-GTCTCGGATAGTGGGCTCTG-3′ (forward) and 5′-TGCTGGCCTTT CTTTCATTT-3′ (reverse) for STAT1; and 5′-TGACGGGGTCACCCAC ACTGTGCCCATCTA-3′ (forward) and 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′ (reverse) for β-actin. PCR products were detected by electrophoresis and confirmed by sequence.
For real-time PCR analysis of B7-H1 mRNA, total RNAs were extracted using TRIzol reagent (Ambion) and cDNA (from 200 ng RNA) was synthesized with the TaqMan reverse transcription kit (Applied Biosystems). The real-time PCR was conducted in triplicate using the SYBR Green PCR master mix (Applied Biosystems) on an Applied Biosystems 7500 FAST real-time cycler. The primers were as follows: 5′-GGTGCCGACTACAAGCGAAT-3′ (forward) and 5′-GGTGACTGGATCCACAACCAA-3′ (reverse) for human B7-H1; and 5′-TGCACCACCAACTGCTTAGC-3′ (forward) and 5′-GGCATGGACTGTGGTCATGAG-3′ (reverse) for human GAPDH. The threshold cycle (Ct) values were analyzed using the comparative Ct (ΔΔCt) method. The amount of target was obtained by normalizing to the endogenous reference (GAPDH) and relative to a (untreated cell) control as previously reported (22).
For real-time PCR analysis of miR-513, total RNA was isolated from cells with the mirVana miRNA isolation kit (Ambion). An amount of 0.25 μg of total RNA was reverse transcribed by using the TaqMan microRNA reverse transcription kit (Applied Biosystems). Comparative real-time PCR was performed by using the TaqMan universal PCR master mix (Applied Biosystems) on the Applied Biosystems 7500 FAST real-time PCR System. Specific primers and probes for mature miR-513 and small nuclear RNA RNU6B (endogenous reference) were obtained from Applied Biosystems. All reactions were run in triplicate. The amount of miR-513 was obtained by normalizing to small nuclear RNA RNU6B and relative to a (untreated cell) control as reported by others (23, 24).
miR-513 precursor and antisense oligonucleotide
To manipulate cellular abundance of miR-513 in H69 cells, we used an antisense oligonucleotide specific to miR-513 to inhibit its function and the miR-513 precursor to increase miR-513 expression. For experiments, H69 cells were grown to 80% confluence and treated with miR-513 antisense 2-methoxy oligonucleotide (Ambion) or the miR-513 precursor (Ambion) using the Lipofectamine 2000 agent (Invitrogen). Nonspecific antisense and precursor (Ambion) were used as control.
Luciferase reporter constructs and luciferase assay
Complementary 34-bp DNA oligonucleotides containing the putative miR-513 target site in the 3′-UTR of human B7-H1 and the flanking SpeI and HindIII restriction enzyme digestion sites (antisense: 5′-ctagGCCTGAGGGGCTCATCGACGCCTGTGACAG-3′; sense: 5′-agctCTGTCACAGGCGTCGATGAGCCCCTCAGGC-3′) were synthesized and cloned into the multiple cloning sites of the pMIR-REPORT Luciferase vector (Ambion). The sense and antisense strands of the oligonucleotides were annealed by adding 200 ng of each oligonucleotide to 100 μl of 1 × SSC (trisodium citrate and sodium chloride (pH 7.0)) and incubated at 90°C for 10 min and then at 37°C for 1 h. The annealed oligonucleotides were ligated into the SpeI/HindIII sites of the pMIR-REPORT luciferase vector (Ambion). In this vector, the posttranscriptional regulation of luciferase was potentially regulated by miRNA interactions with the B7-H1 3′-UTR. Another pMIR-REPORT luciferase construct containing mutant 3′-UTR (GTGAC to TGGAC) was also generated as a control. We then transfected cholangiocytes with each reporter construct as well as miR-513 antisense oligonucleotide or precursor using the Lipofectamine 2000 agent, followed by assessment of luciferase activity 24 h after transfection. Luciferase activity was normalized to the control β-galactosidase level as previously reported (18).
Coculture of H69 cells with Jurkat cells
H69 and Jurkat cells were cocultured as previously reported (25). Briefly, H69 cells were seeded at 2 × 105 cells/well in 24-well plates and exposed to IFN-γ for 24 h in the presence or absence of a neutralizing Ab to B7-H1 (clone 5H1-A3; 5 μg/ml) (4). Some H69 cells were transfected with the miR-513 precursor or the nonspecific control precursor for 48 h before IFN-γ stimulation. H69 cells were then cocultured with Jurkat cells (2 × 105) in the culture medium containing 20 nM PMA (Sigma-Aldrich) for 24 h as previously reported (26) in the presence or absence of a neutralizing Ab to Fas (clone ZB4; 5 μg/ml) (Assay Designs) (27). Jurkat cells were then collected from the medium separating them from H69 cells, which were attached to the plates. Apoptotic cell death in Jurkat cells was measured by 4′,6-diamidino-2-phenylindole (DAPI) staining as previously reported (25).
Statistical analysis
Groups of data were compared using the Student’s t test. p < 0.05 was considered to represent statistical significance.
Results
Posttranscriptional suppression of B7-H1 exists in human cholangiocytes and IFN-γ induces cholangiocyte B7-H1 protein expression
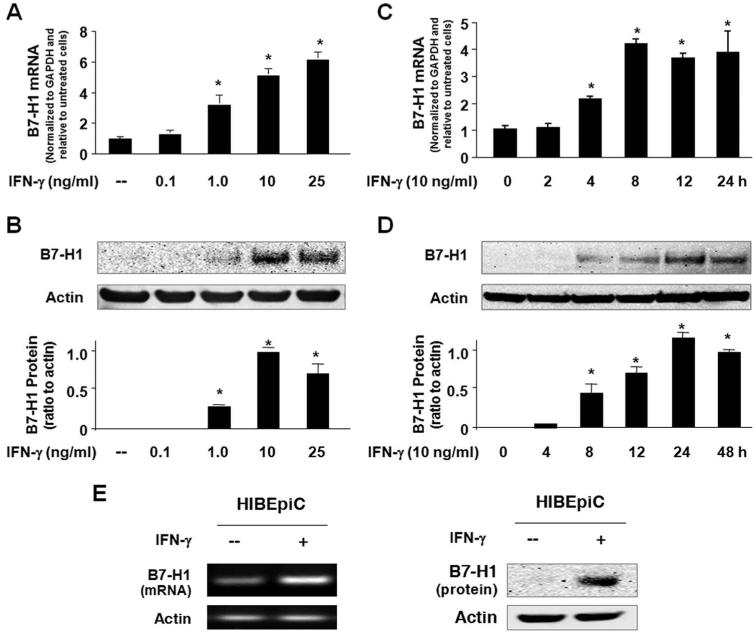
We first assessed IFN-γ-induced B7-H1 expression in H69 cells. A considerable amount of B7-H1 mRNA was detected by real-time PCR in nonstimulated H69 cells and a dose-dependent increase of B7-H1 mRNA levels was observed in cells after exposure to IFN-γ for 8 h (Fig. 1A). In contrast, B7-H1 protein was barely detectable in nonstimulated cells by Western blotting (Fig. 1B) or immunofluorescent staining using multiple Abs from different resources (data not shown), suggesting posttranscriptional suppression, a phenomenon reported previously in other epithelial cells and T cells (4, 5). Accordingly, B7-H1 protein became detectable in cells after exposure to 1 ng/ml IFN-γ for 24 h, and maximal B7-H1 protein expression was observed in cells exposed to 10 ng/ml IFN-γ (Fig. 1B). A time-dependent increase of B7-H1, at both message and protein levels, was detected in H69 cells exposed to 10 ng/ml IFN-γ (Fig. 1, C and D). Increase of B7-H1 mRNA started at 4 h after IFN-γ stimulation, reached a peak at 8 h, and then leveled off up to 24 h. Expression at the protein level started at 8 h and increased up to 48 h. To further confirm B7-H1 expression in human cholangiocytes in response to IFN-γ, we tested B7-H1 expression in HIBEpiC cells, nonimmortalized isolated primary human cholangiocytes. Consistent with what we found in H69 cells, we detected expression of B7-H1 mRNA but not B7-H1 protein (Fig. 1E). When HIBEpiC cells were exposed to 10 ng/ml IFN-γ for 8 h, an increase of B7-H1 mRNA was detected. B7-H1 protein was detected in cells after exposure to IFN-γ for 24 h (Fig. 1E). Taken together, our above data suggest that posttranscriptional suppression of B7-H1 expression exists in human cholangiocytes. IFN-γ increases B7-H1 transcription and induces cholangiocyte B7-H1 protein expression.
FIGURE 1.
Posttranscriptional suppression of B7-H1 exists in human cholangiocytes and IFN-γ induces cholangiocyte B7-H1 protein expression. A and B, Dose-dependent expression of B7-H1 at the message (A) and protein (B) levels in H69 cells following IFN-γ stimulation. H69 cells were exposed to culture medium with various doses of IFN-γ (0, 0.1, 1.0, 10, and 25 ng/ml) for 8 h (for real-time PCR) or 24 h (for Western blotting). A representative Western blot from three independent experiments is shown in B. Actin was blotted as a loading control. Densitometric levels of B7-H1 signals were quantified and expressed as the ratio to actin. C and D, Time-dependent expression of B7-H1 expression in H69 cells induced by IFN-γ. Cells were exposed to IFN-γ (10 ng/ml) for 2-48 h followed by real-time PCR (C) and Western blotting (D) for B7-H1. E, Expression of B7-H1 in HIBEpiC cells upon IFN-γ stimulation. HIBEpiC cells were exposed to culture medium with or without IFN-γ (10 ng/ml) for 8 h (for RT-PCR) or 24 h (for Western blotting). *, p < 0.05, vs the nonstimulated control.
IFN-γ alters miRNA expression profile in cholangiocytes and decreases miR-513 expression in a STAT1-dependent manner
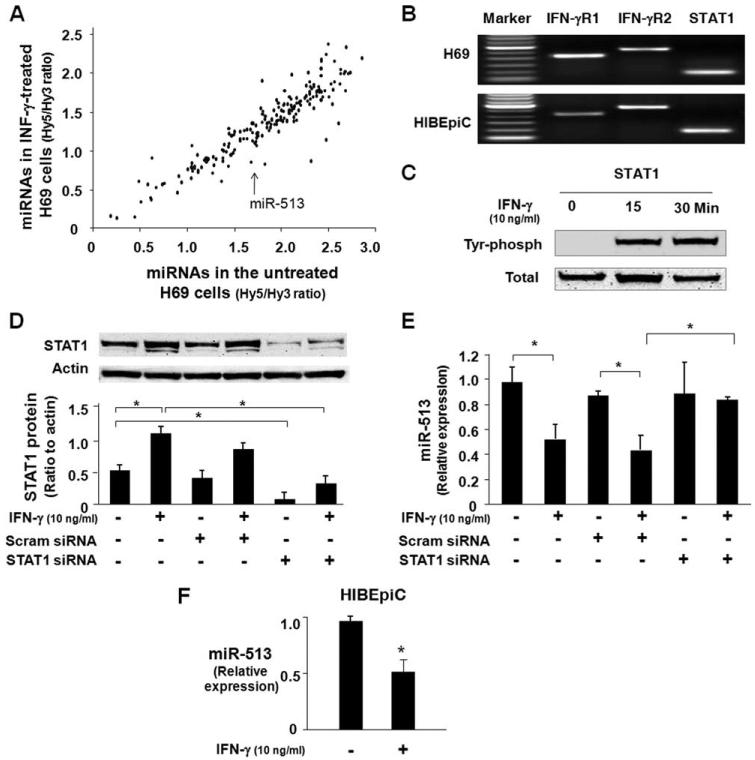
Using the miRCURY LNA array analysis service provided and performed by Exiqon, we detected expression of 206 miRNAs in H69 cells (Fig. 2A). A total of 76 miRNAs were significantly down-regulated in H69 cells after exposure to IFN-γ (10 ng/ml) for 8 h, including miR-513 (Fig. 2A and Table 1). We also determined that miR-29b-1* was significantly up-regulated after IFN-γ stimulation. No significant change for other miRNAs was detected between nonstimulated and IFN-γ-stimulated cells (data not shown). All microarray data reported in this article were described in accordance with MIAME (minimum information about a microarray experiment; Microarray and Gene Expression Data (MGED) Society) guidelines and were deposited at ArrayExpress under accession no. E-MEXP-1906 (www.ebi. ac.uk/microarray-as/ae/).
FIGURE 2.
IFN-γ alters miRNA expression profile in cholangiocytes and decreases miR-513 expression in cholangiocytes in a STAT1-dependent manner. A, Microarray analysis of miRNA expression in H69 cells after IFN-γ stimulation for 8 h. Data were presented as the mean values of the mean Hy5/Hy3 ratios from three independent experiments. Each dot represents one miRNA. B, Expression of IFN-γ receptors and STAT1 in cholangiocytes. The mRNAs of two IFN-γ receptors (IFN-γR1 and IFN-γR2) and STAT1 were detected in both H69 cells and HIBEpiC cells by RT-PCR. C, Activation of STAT1 via tyrosine phosphorylation in cholangiocytes induced by IFN-γ. H69 cells were exposed to IFN-γ (10 ng/ml) for 15 or 30 min and tyrosine phosphorylation of STAT1 was assessed by Western blotting using Abs recognizing the tyrosine-phosphorylated (Tyr phosph) form of STAT1. Total STAT1 was also blotted as a control. D, Knockdown of STAT1 expression in H69 cells by siRNA gene silencing. H69 cells were treated with a siRNA to STAT1 for 48 h with an additional incubation for 8 h in the absence or presence of IFN-γ (10 ng/ml) followed by Western blotting for STAT1. A scrambled control siRNA (Scram siRNA) was used as the control. E, Knockdown of STAT1 blocked IFN-γ-induced decrease of miR-513 expression. H69 cells were treated with either the STAT1 siRNA or scrambled control siRNA for 48 h with an additional incubation for 8 h in the absence or presence of IFN-γ (10 ng/ml). Total RNA was isolated and the expression of miR-513 was quantified by real-time PCR. Data are representative of three independent experiments. F, IFN-γ decreases miR-513 expression in HIBEpiC cells. Cells were exposed to IFN-γ (10 ng/ml) for 8 h followed by real-time PCR for miR-513. *, p < 0.05, vs the nonstimulated control (in F) or as indicated (in D and E).
Table I.
miRNAs that are significantly changed in H69 cells following IFN-γ stimulation
| Log2 (Hy5/Hy3) Ratios |
|||
|---|---|---|---|
| miRNAs | Control | IFN-γ | p |
| hsa-let-7e | 1.226 ± 0.031 | 0.744 ± 0.215 | 0.025 |
| hsa-let-7f | 0.998 ± 0.078 | 0.508 ± 0.178 | 0.021 |
| hsa-let-7i | 0.963 ± 0.049 | 0.531 ± 0.245 | 0.049 |
| hsa-miR-1 | 0.324 ± 0.291 | -0.489 ± 0.128 | 0.037 |
| hsa-miR-100 | 1.420 ± 0.177 | 0.811 ± 0.225 | 0.041 |
| hsa-miR-106b | 1.391 ± 0.071 | 0.887 ± 0.218 | 0.028 |
| hsa-miR-107 | 0.927 ± 0.084 | 0.476 ± 0.225 | 0.044 |
| hsa-miR-125a-5p | 1.109 ± 0.083 | 0.683 ± 0.223 | 0.049 |
| hsa-miR-126 | 0.362 ± 0.057 | -0.318 ± 0.351 | 0.037 |
| hsa-miR-129* | 0.758 ± 0.114 | 0.178 ± 0.026 | 0.007 |
| hsa-miR-138 | 1.139 ± 0.096 | 0.665 ± 0.166 | 0.025 |
| hsa-miR-148b | 0.509 ± 0.063 | -0.035 ± 0.308 | 0.049 |
| hsa-miR-15a | 1.204 ± 0.057 | 0.813 ± 0.145 | 0.021 |
| hsa-miR-15b | 1.336 ± 0.112 | 0.885 ± 0.181 | 0.038 |
| hsa-miR-16 | 1.422 ± 0.064 | 1.009 ± 0.196 | 0.036 |
| hsa-miR-182 | 0.792 ± 0.112 | 0.153 ± 0.218 | 0.021 |
| hsa-miR-183 | 1.297 ± 0.221 | 0.149 ± 0.496 | 0.034 |
| hsa-miR-185 | 1.091 ± 0.126 | 0.517 ± 0.103 | 0.013 |
| hsa-miR-186 | -0.708 ± 0.030 | -1.354 ± 0.376 | 0.048 |
| hsa-miR-193b | 1.062 ± 0.088 | 0.550 ± 0.248 | 0.039 |
| hsa-miR-195 | 0.965 ± 0.087 | 0.454 ± 0.261 | 0.044 |
| hsa-miR-196a* | 0.518 ± 0.072 | -0.317 ± 0.171 | 0.004 |
| hsa-miR-197 | 0.520 ± 0.063 | 0.068 ± 0.184 | 0.025 |
| hsa-miR-19a | 1.266 ± 0.054 | 0.827 ± 0.213 | 0.035 |
| hsa-miR-19b | 1.280 ± 0.163 | 0.757 ± 0.154 | 0.037 |
| hsa-miR-20a | 1.448 ± 0.025 | 0.995 ± 0.244 | 0.040 |
| hsa-miR-214 | 0.269 ± 0.100 | -0.167 ± 0.094 | 0.016 |
| hsa-miR-22 | 1.278 ± 0.122 | 0.803 ± 0.155 | 0.030 |
| hsa-miR-23b | 1.204 ± 0.009 | 0.671 ± 0.228 | 0.021 |
| hsa-miR-24 | 1.093 ± 0.058 | 0.622 ± 0.146 | 0.013 |
| hsa-miR-24-1* | 0.799 ± 0.064 | 0.085 ± 0.227 | 0.012 |
| hsa-miR-28-5p | 1.186 ± 0.098 | 0.699 ± 0.243 | 0.046 |
| hsa-miR-29b-1* | -0.664 ± 0.082 | -0.155 ± 0.205 | 0.026 |
| hsa-miR-300 | 0.811 ± 0.015 | 0.243 ± 0.133 | 0.004 |
| hsa-miR-302d* | 1.011 ± 0.082 | 0.456 ± 0.072 | 0.004 |
| hsa-miR-30b | 1.258 ± 0.067 | 0.810 ± 0.206 | 0.033 |
| hsa-miR-30b* | 0.887 ± 0.065 | 0.371 ± 0.198 | 0.021 |
| hsa-miR-30c-2* | 1.106 ± 0.081 | 0.554 ± 0.135 | 0.010 |
| hsa-miR-30d | 0.801 ± 0.124 | 0.296 ± 0.162 | 0.028 |
| hsa-miR-423-3p | 1.131 ± 0.033 | 0.675 ± 0.253 | 0.044 |
| hsa-miR-452 | 0.389 ± 0.057 | -0.191 ± 0.302 | 0.039 |
| hsa-miR-491-3p | 0.798 ± 0.088 | 0.286 ± 0.115 | 0.011 |
| hsa-miR-513 | 0.746 ± 0.134 | -0.262 ± 0.463 | 0.032 |
| hsa-miR-518c* | 0.799 ± 0.132 | 0.219 ± 0.090 | 0.013 |
| hsa-miR-549 | 1.106 ± 0.141 | 0.471 ± 0.022 | 0.009 |
| hsa-miR-550 | 1.042 ± 0.142 | 0.465 ± 0.232 | 0.038 |
| hsa-miR-551b | 1.323 ± 0.123 | 0.755 ± 0.124 | 0.015 |
| hsa-miR-574-5p | 0.777 ± 0.131 | 0.138 ± 0.027 | 0.007 |
| hsa-miR-576-3p | 0.980 ± 0.118 | 0.315 ± 0.172 | 0.013 |
| hsa-miR-583 | 0.557 ± 0.111 | -0.079 ± 0.036 | 0.005 |
| hsa-miR-600 | 0.439 ± 0.221 | -0.768 ± 0.484 | 0.028 |
| hsa-miR-620 | 1.227 ± 0.145 | 0.634 ± 0.008 | 0.012 |
| hsa-miR-620 | 1.087 ± 0.172 | 0.462 ± 0.001 | 0.016 |
| hsa-miR-634 | 0.641 ± 0.087 | 0.197 ± 0.179 | 0.030 |
| hsa-miR-642 | 0.468 ± 0.131 | -0.138 ± 0.081 | 0.011 |
| hsa-miR-658 | 1.044 ± 0.118 | 0.460 ± 0.055 | 0.008 |
| hsa-miR-665 | 1.092 ± 0.098 | 0.637 ± 0.035 | 0.009 |
| hsa-miR-765 | 0.895 ± 0.173 | 0.333 ± 0.001 | 0.022 |
| hsa-miR-886-5p | 1.693 ± 0.181 | 0.639 ± 0.533 | 0.043 |
| hsa-miR-921 | 1.251 ± 0.060 | 0.814 ± 0.126 | 0.012 |
| hsa-miR-93 | 1.125 ± 0.072 | 0.594 ± 0.255 | 0.035 |
| hsa-miR-939 | 0.947 ± 0.082 | 0.415 ± 0.061 | 0.005 |
| hsa-miR-96 | 0.844 ± 0.108 | 0.252 ± 0.203 | 0.021 |
| miRPlus_17869 | 1.207 ± 0.057 | 0.720 ± 0.232 | 0.033 |
| miRPlus_42487 | 0.973 ± 0.108 | 0.465 ± 0.214 | 0.035 |
| miRPlus_42530 | 0.246 ± 0.078 | -0.200 ± 0.199 | 0.034 |
| hsa-miR-30e | 1.247 ± 0.073 | 0.758 ± 0.234 | 0.036 |
| hsa-miR-30e* | 0.164 ± 0.097 | -0.491 ± 0.313 | 0.036 |
| hsa-miR-31* | 0.996 ± 0.135 | 0.473 ± 0.180 | 0.032 |
| hsa-miR-32* | 0.922 ± 0.024 | 0.415 ± 0.015 | 0.000 |
| hsa-miR-339-5p | 0.882 ± 0.098 | 0.428 ± 0.140 | 0.022 |
| hsa-miR-340 | 0.964 ± 0.115 | 0.414 ± 0.222 | 0.032 |
| hsa-miR-342-3p | 0.785 ± 0.060 | 0.109 ± 0.170 | 0.007 |
| hsa-miR-34a | 1.179 ± 0.112 | 0.769 ± 0.110 | 0.027 |
| hsa-miR-34b | 1.344 ± 0.121 | 0.676 ± 0.020 | 0.005 |
| hsa-miR-374a | 1.350 ± 0.072 | 0.831 ± 0.206 | 0.023 |
| hsa-miR-374b* | -0.152 ± 0.072 | -1.138 ± 0.123 | 0.001 |
Data represent the log2 (Hy5/Hy3) ratios from nonstimulated and IFN-γ-stimulated (10 ng/ml) samples by using the miRCURY LNA Array (version 10.0). A total of 76 miRNAs were down-regulated and miR-29b-1* was up-regulated in H69 cells following IFN-γ stimulation for 8 h (p < 0.05).
Key elements for IFN-γ signaling pathways include membrane IFN-γ receptors and activation of STAT transcription factors via tyrosine phosphorylation (28). Using RT-PCR analysis, we detected the mRNAs of IFN-γR1, IFN-γR2, and STAT1 in both H69 and HIBEpiC cells (Fig. 2B). IFN-γ stimulation induced a significant increase of STAT1 tyrosine phosphorylation in H69 cells (Fig. 2C). To test whether STAT1 is involved in IFN-γ-altered miRNA expression, we used a specific siRNA (Cell Signaling) to knock down STAT1 and assessed its effects on IFN-γ-associated expression of miR-513. Cells were transfected with STAT1 siRNA for 48 h followed by an additional incubation for 8 h with or without IFN-γ (10 ng/ml). As shown in Fig. 2D, STAT1 siRNA induced a significant decrease of STAT1 protein expression in both nontreated and IFN-γ-treated cells. A scrambled siRNA did not show any effects on cellular STAT1 level. Consistent with our microarray analysis results, a significant decrease of miR-513 level was detected in H69 cells exposed to IFN-γ for 8 h as assessed by real-time PCR (Fig. 2E). H69 cells treated with the scrambled siRNA showed a similar decrease of miR-513 expression following IFN-γ stimulation. In contrast, no IFN-γ-associated decrease of miR-513 was found in cells treated with the STAT1 siRNA (Fig. 2E). No change of miR-513 expression was identified between cells treated with STAT1 siRNA alone and non-IFN-γ-stimulated control cells or cells treated with the scrambled siRNA (Fig. 2E). Moreover, decrease of miR-513 was also detected in HIBEpiC cells after exposure to IFN-γ for 8 h (Fig. 2F).
miR-513 targets a binding site in the B7-H1 3′-UTR
In mammal cells, miRNAs target messenger RNAs via base pair complementarity with the 3′-UTR of target mRNAs, leading to either mRNA cleavage or translational suppression (14, 15). Of those IFN-γ-responsive miRNAs expressed in H69 cells, we identified a considerable complementarity within the seed region of miR-513 and the 3′-UTR of B7-H1 (Fig. 3A) using the algorithms in TargetScan 4.2 (http://www.Targetscan.org/) (29).
FIGURE 3.
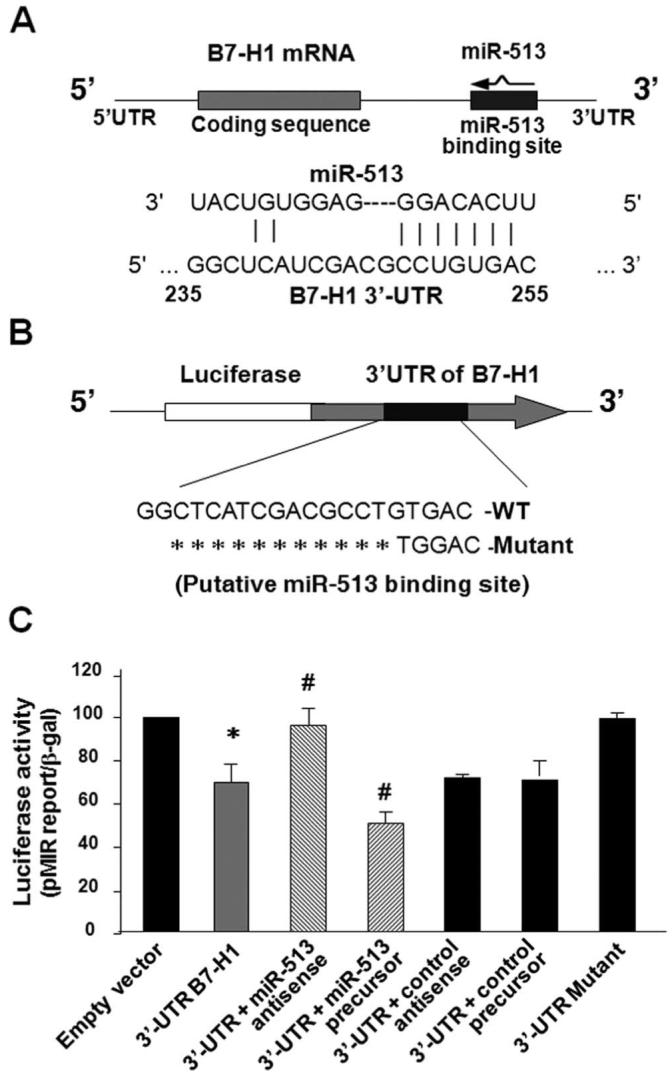
miR-513 targets a potential binding site in the B7-H1 3′-UTR resulting in translational suppression. A, The schematic of B7-H1 mRNA shows a potential binding site in the B7-H1 3′-UTR for miR-513. B, The complementary miR-513-binding site in the B7-H1 3′-UTR was inserted downstream of a luciferase reporter on the pMIR-Report plasmid. A control plasmid with the mutant 3′-UTR sequence was also generated. WT, Wild type. C, Targeting of B7-H1 3′-UTR by miR-513 results in translational suppression. H69 cells were transfected with the reporter constructs simultaneously with or without the miR-513 antisense oligonucleotide or the miR-513 precursor. After 24 h, cells were harvested and luciferase activities were measured and normalized to the control β-galactosidase level. A scrambled antisense oligonucleotide and a nonspecific precursor were used as the controls. These data are representative of three independent experiments. *, p < 0.05, vs the empty vector; #, p < 0.05, vs the B7-H1 3′-UTR reporter construct.
To explore the potential targeting of B7-H1 mRNA by miR-513, we first generated a pMIR-REPORT luciferase construct that contains the 3′-UTR of B7-H1 with the putative miR-513 binding site (Fig. 3B). In addition, another pMIR-REPORT luciferase construct containing B7-H1 mRNA 3′-UTR with a mutation at the putative miR-513 binding site (GTGAC to TGGAC) was generated as a control (Fig. 3B). We then transfected cells with each reporter construct followed by assessment of luciferase activity 24 h after transfection. As shown in Fig. 3C, a significant decrease of luciferase activity was detected in cells transfected with the B7-H1 3′-UTR construct compared with the empty vector control, suggesting endogenous translational repression of the construct with the B7-H1 3′-UTR. Using a well-established approach (18, 30) with specific miR-513 antisense and miR-513 precursor to inhibit or increase miR-513 abundance, respectively, we further tested effects of miR-513 on the translation of the construct with the B7-H1 3′-UTR. The miR-513 antisense significantly increased B7-H1 3′-UTR-associated luciferase reporter translation (Fig. 3C). In contrast, miR-513 precursor markedly decreased the luciferase translation (Fig. 3C). In the controls, nonspecific antisense and precursor had no effect on 3′-UTR-associated luciferase translation and no difference in luciferase activity was found in cells transfected with the 3′-UTR mutant compared with the empty vector control (Fig. 3C). These results suggest that miR-513 can target the B7-H1 3′-UTR and, subsequently, cause translational repression in cholangiocytes.
miR-513 antisense transfection induces B7-H1 protein expression in cholangiocytes
To test whether miR-513-mediated translational repression of B7-H1 is directly relevant to B7-H1 protein expression in cholangiocytes, we treated H69 cells with the miR-513 antisense or a control nonspecific antisense for 72 h and then assessed B7-H1 protein expression by Western blotting. Consistent with our previous results, B7-H1 protein was not detectable in nontreated cells. Importantly, the miR-513 antisense induced a dose-dependent expression of B7-H1 protein in H69 cells (Fig. 4A). In contrast, no B7-H1 protein was detected in cells treated with the control antisense. Moreover, no significant change in B7-H1 mRNA levels was found between the control cells and cells treated with miR-513 antisense (Fig. 4B), suggesting that miR-513 antisense does not affect cellular B7-H1 mRNA levels. Taken together, our data indicate that miR-513 mediates translational repression of B7-H1, a process that may account for the posttranscriptional suppression of B7-H1 in resting human cholangiocytes.
FIGURE 4.
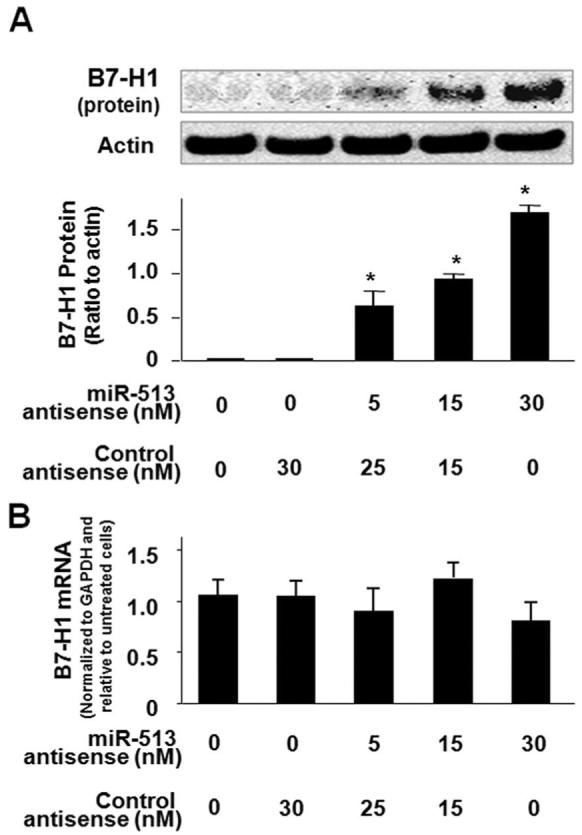
miR-513 antisense transfection induces B7-H1 protein expression in H69 cells. A, Transfection of miR-513 antisense induces B7-H1 protein expression. H69 cells were treated with an antisense oligonucleotide to miR-513 or a nonspecific control antisense for 72 h followed by Western blotting for B7-H1. A representative Western blot from three independent experiments and densitometric levels of B7-H1 signals are shown. *, p < 0.05, vs the nonantisense-treated control. B, miR-513 antisense transfection does not affect B7-H1 mRNA levels. H69 cells were exposed to miR-513 antisense or a nonspecific control antisense for 72 h followed by real-time PCR analysis for B7-H1 mRNA.
Transfection of miR-513 precursor reduces IFN-γ-induced B7-H1 protein expression in cholangiocytes
To test whether relief of miR-513-mediated B7-H1 translational repression is involved in IFN-γ-induced B7-H1 protein expression in cholangiocytes, we first transfected H69 cells with various doses of miR-513 precursor for 48 h and then exposed those cells to IFN-γ for 24 h, followed by Western blot analysis for B7-H1 protein. As shown in Fig. 5A, miR-513 precursor significantly blocked IFN-γ-induced B7-H1 protein expression in H69 cells in a dosedependent manner. In contrast, a control RNA sequence did not show any effects. No significant change in B7-H1 mRNA levels was found in IFN-γ-stimulated cells treated with miR-513 precursor compared with IFN-γ-stimulated cells treated with the control precursor (Fig. 5B). Moreover, no significant change in B7-H1 mRNA levels was found between the control cells and cells treated with miR-513 precursor for 48 h in the absence of IFN-γ (Fig. 5C). Coupled with the down-regulation of cellular miR-513 in response to IFN-γ, the above data suggest that relief of miR-513-mediated translational repression of B7-H1 may be involved in IFN-γ-induced B7-H1 protein expression in cholangiocytes.
FIGURE 5.
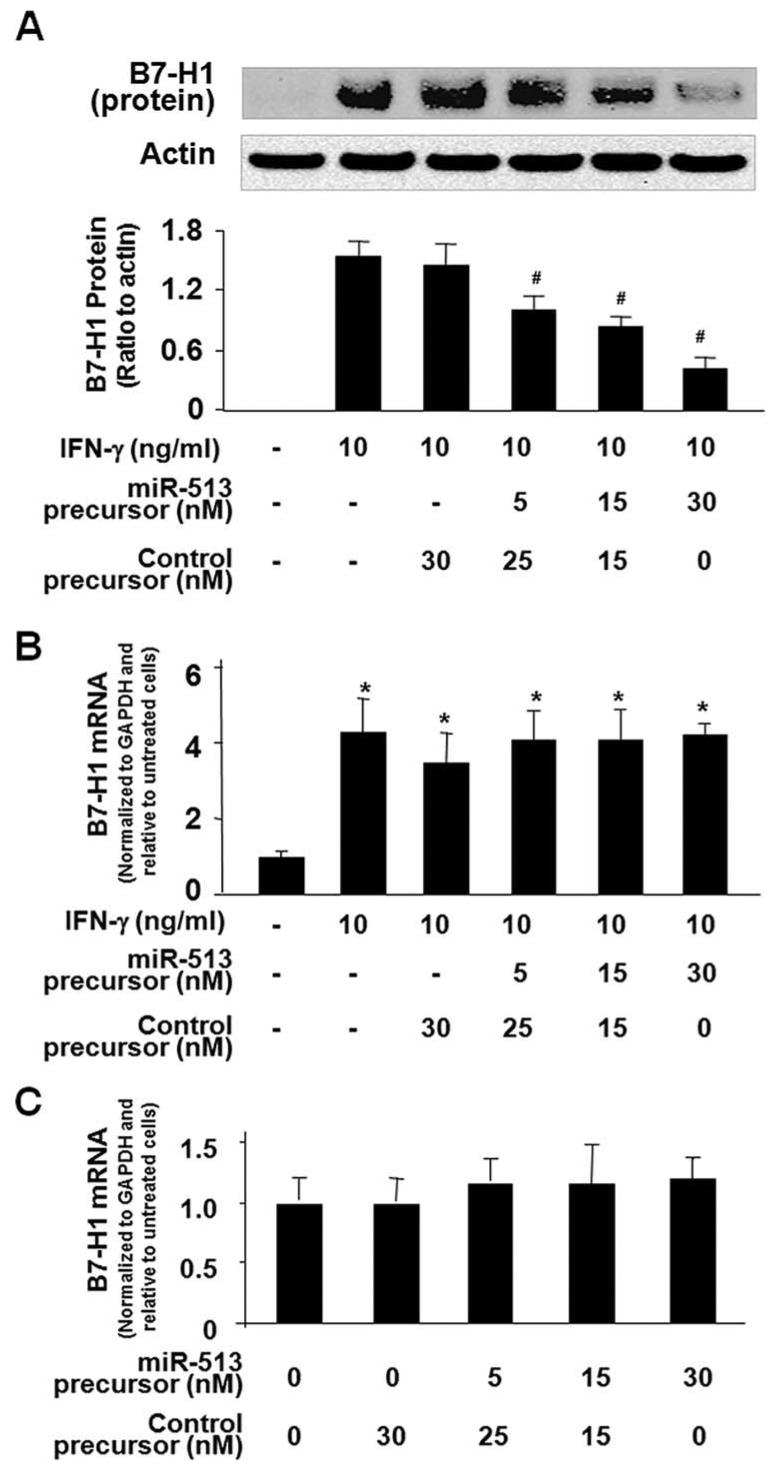
Transfection of miR-513 precursor reduces IFN-γ-induced B7-H1 protein expression in H69 cells. A, miR-513 precursor reduces IFN-γ-induced B7-H1 protein expression. H69 cells were transfected with miR-513 precursor or a control nonspecific precursor for 48 h and then exposed to IFN-γ (10 ng/ml) for 24 h followed by Western blotting for B7-H1. A representative Western blot from three independent experiments and densitometric levels of B7-H1 signals are shown. #, p < 0.05, vs IFN-γ-treated alone. B, miR-513 precursor transfection does not affect IFN-γ-stimulated B7-H1 mRNA expression. H69 cells were transfected with miR-513 precursor or a control nonspecific precursor for 48 h and then exposed to IFN-γ (10 ng/ml) for 24 h followed by real-time PCR analysis for B7-H1 mRNA. *, p < 0.05, vs non-IFN-γ-stimulated control. C, miR-513 precursor transfection does not affect B7-H1 mRNA expression in nonstimulated cells. H69 cells were transfected with miR-513 precursor or a control nonspecific precursor for 48 h in the absence of IFN-γ followed by B7-H1 mRNA real-time PCR analysis.
miR-513 influences B7-H1-associated apoptotic cell death in cocultured Jurkat cells
B7-H1 has been reported to induce apoptotic cell death in activated T cells (4, 6, 10). We then tested apoptotic cell death in Jurkat cells when cocultured with H69 cells stimulated with IFN-γ in the presence or absence of neutralizing Abs to B7-H1 or Fas. Because nonstimulated Jurkat cells do not express PD-1 (26), the receptor for B7-H1, we used 20 nM PMA (Sigma-Aldrich) to induce PD-1 expression in Jurkat cells as previously reported (26). A significant increase of apoptosis was detected in Jurkat cells after coculture with IFN-γ-stimulated H69 cells (Fig. 6). In addition, apoptosis in cocultured Jurkat cells was partially blocked by pretreatment of H69 cells with a B7-H1-neutralizing Ab or miR-513 precursor (Fig. 6). Because activation of Jurkat cells was induced by incubation with PMA, not by allogeneic reaction, the potential histocompatibility between two cell types should not be a factor for their coculture. In addition, a neutralizing Ab to Fas did not show significant inhibitory effects on the associated apoptosis (Fig. 6), suggesting that the Fas/Fas ligand pathway may not be involved in the apoptosis in the cocultured Jurkat cells.
FIGURE 6.
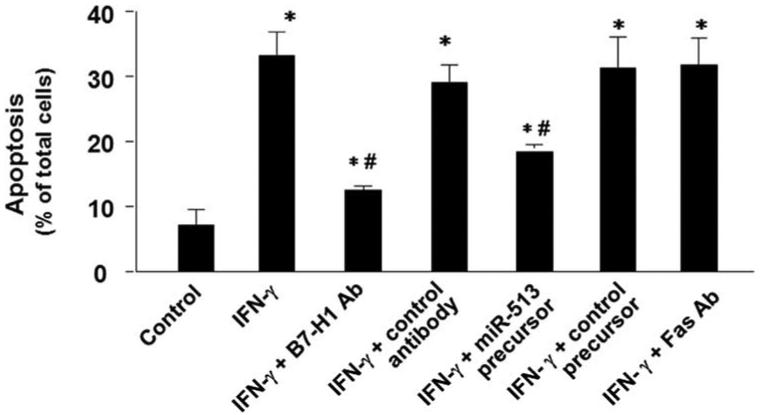
miR-513 influences B7-H1-associated apoptotic cell death in cocultured Jurkat cells. H69 cells were first exposed to IFN-γ for 24 h in the presence or absence of a neutralizing Ab to B7-H1. Some cells were transfected with miR-513 precursor or a control precursor for 48 h before exposure to IFN-γ. H69 cells were then cocultured with Jurkat cells in the presence of PMA with or without a neutralizing Ab to Fas for 24 h. Jurkat cells were separated from H69 cells and harvested for apoptosis assay by DAPI staining. These data are representative of three independent experiments. Ab; *, p < 0.05, vs Jurkat cells cocultured with non-IFN-γ-stimulated H69 cells as the control; #, p < 0.05, vs Jurkat cells cocultured with IFN-γ-stimulated H69 cells without Ab or precursor treatment.
Discussion
The key findings in this report are the following: 1) posttranscriptional repression of B7-H1 exists in human cholangiocytes; 2) IFN-γ alters cholangiocyte miRNA expression profile and miR-513, one of the IFN-γ-down-regulated miRNAs, targets B7-H1, resulting in translational repression; and 3) relief of miR-513-mediated translational repression may be involved in IFN-γ-induced expression of B7-H1 expression in cholangiocytes and consequently influence cholangiocyte-T cell interactions. These data indicate that miR-513 targets B7-H1 and is involved in IFN-γ-induced B7-H1 expression in cholangiocytes, suggesting a role for miRNA-mediated gene expression in the regulation of cholangiocyte response to IFN-γ.
Expression of B7-H1 mRNA, but not at the protein level, has been previously demonstrated in many cell types (4, 5). Inflammatory cytokines and chemokines, such as IFN-γ and TNF-α, are potent activators for inducing B7-H1 protein expression in T cells, B cells, endothelial cells, and epithelial cells (6). Consistently, we detected B7-H1 mRNA, but not B7-H1 protein, in both SV40transformed human cholangiocytes and isolated human cholangiocytes, suggesting posttranscriptional suppression of B7-H1 protein expression in resting human cholangiocytes. We also demonstrated that IFN-γ increases expression of B7-H1 at both the message and protein levels in cholangiocytes, suggesting an induced expression of B7-H1 in response to IFN-γ stimulation. The mRNA expression of B7-H1 was previously demonstrated in several other human cholangiocyte cell lines, and B7-H1 protein was detected in those cells after treatment with IFN-γ, TNF-α, and LPS (13).
IFN-γ triggers both gene trans-activation and trans-suppression in stimulated cells (31-34). In the present study, we found that a total of 76 miRNAs were down-regulated and one miRNA was up-regulated in cholangiocytes following IFN-γ stimulation. Similarly, differential expression of cellular miRNAs induced by IFN was previously reported in hepatocytes (35). Key elements for IFN-γ signaling pathways include membrane IFN-γ receptors and activation of downstream signals, primarily the activation of JAK and STAT transcription factors via tyrosine phosphorylation (28, 36). Using RT-PCR analysis, we detected the mRNAs of two IFN-γ receptors, IFN-γR1 and IFN-γR2, in both H69 and HIBEpiC cells. IFN-γ stimulation induced a time-dependent tyrosine phosphorylation of STAT1 and, interestingly, knockdown of STAT1 via siRNA silencing could block IFN-γ-induced decrease of miR-513 expression. These observations suggest to us that STAT1 may be involved in IFN-γ-induced miR-513 down-regulation in cholangiocytes. Similarly, other transcription factors, including c-Myc and NF-κB, have recently been implicated in the regulation of miRNA genes (18, 37). Whether IFN-γ alters other miRNA expression in cholangiocytes via the STAT signal pathway and whether IFN-γ may induce a global decrease of miRNA expression require further investigation.
Several findings from this study support the idea that miR-513 mediates B7-H1 translational repression and may be involved in the posttranscriptional suppression in nonstimulated resting cholangiocytes. A significant decrease of luciferase activity was detected in cells transfected with a pMIR-REPORT luciferase construct that contains the putative miR-513 binding site in the 3′-UTR of B7-H1 compared with cells transfected with the control empty vector, suggesting that a 3′-UTR-associated B7-H1 translational inhibition exists in nonstimulated cholangiocytes. Whereas a reasonable level of miR-513 was detected by real-time PCR analysis in cells under resting conditions, miR-513 precursor and antisense oligonucleotide specific to miR-513 caused reciprocal alterations in B7-H1 3′-UTR-associated luciferase reporter translation. Thus, endogenous expression of miR-513 may be involved in the 3′-UTR-associated B7-H1 translational inhibition in resting cholangiocytes. Indeed, treatment of cells at resting condition with miR-513 antisense could induce a significant cellular expression of B7-H1 protein. No changes in B7-H1 mRNA levels were detected in cells treated with miR-513 antisense, suggesting that miR-513 antisense may not directly affect transcription or degradation of B7-H1 mRNA.
A potential role for miR-513-mediated B7-H1 translational repression was further confirmed in cholangiocytes following treatment with IFN-γ. Whereas IFN-γ-induced B7-H1 protein in cholangiocytes, we found that IFN-γ significantly decreased miR-513 expression in cells. Importantly, treatment of cells with miR-513 precursor inhibited IFN-γ-induced B7-H1 protein expression in a dose-dependent manner but did not change B7-H1 mRNA levels. Because a control precursor did not show any inhibitory effect, we speculate that inhibition of induced B7-H1 protein expression by miR-513 precursor is due to the enhanced translational repression via specific binding of miR-513 to the 3′-UTR of B7-H1. Coupled with our observation of increased expression of B7-H1 mRNA following IFN-γ stimulation, our results suggest that IFN-γ-induced B7-H1 protein expression in cholangiocytes involves both an increase of transcription and relief of miR-513-mediated translational repression.
B7-H1 is the ligand for PD-1, a transmembrane receptor of the Ig superfamily expressed on thymocytes, T cells, and B cells (4, 10). Recent studies revealed that B7-H1 possesses dual functions in regulating T cell homeostasis. B7-H1 activates positive signals in naive T cells to stimulate early T cell priming and differentiation (4, 11) but initiates a negative response in activated T cells to inhibit T cell function and survival (4, 11). In this study, we found that B7-H1 expressed on the surface of human cholangiocytes following IFN-γ stimulation induces apoptotic cell death in cocultured activated Jurkat cells. Inhibition of B7-H1 function, either by transfection of cells with miR-513 precursor to suppress B7-H1 expression or by a B7-H1 neutralizing Ab, blocked associated apoptosis in cocultured activated Jurkat cells. Clinical studies had revealed that proinflammatory cytokines such as IFN-γ and TNF-α are associated with the persistent biliary inflammation and cell damage in the portal region in patients with primary biliary cirrhosis (1-3), biliary atresia (38), and primary sclerosing cholangitis (1, 3). Cholangiocyte-associated expression of B7-H1 protein has been reported in patients with inflammatory hepatobiliary disorders including autoimmune hepatitis, primary biliary cirrhosis, and chronic hepatitis type C (12). Thus, expression of B7-H1 in cholangiocytes may be a key element of cholangiocyte immunity and important in the regulation of cholangiocyte-T cell interactions.
In conclusion, our data indicate that miR-513 targets the 3′-UTR of B7-H1, resulting in translational repression. IFN-γ-induced B7-H1 expression in human cholangiocytes involves both increase of transcription and relief of miR-513-mediated translational repression. It will be of interest to extend these studies to determine the mechanisms by which IFN-γ regulates miRNA expression and the role for miRNAs in the regulation of epithelial inflammatory response in general.
Footnotes
This work was supported by National Institutes of Health Grants AI071321 and by the Nebraska Tobacco Settlement Biomedical Research Program LB692 (to X.-M.C.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- miRNA
- microRNA
- DAPI
- 4′,6-diamidino-2-phenylindole
- HIBEpiC
- human intrahepatic biliary epithelial cell
- miR-513
- microRNA-513
- PD-1
- programmed death receptor-1
- siRNA
- small interfering RNA
- UTR
- untranslated region
Disclosures
The authors have no financial conflict of interest.
References
- 1.Lazaridis KN, Strazzabosco M, LaRusso NF. The cholangiopathies: disorders of biliary epithelia. Gastroenterology. 2004;127:1565–1577. doi: 10.1053/j.gastro.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Strazzabosco M, Fabris L, Spirli C. Pathophysiology of cholangiopathies. J. Clin. Gastroenterol. 2005;39:S90–S102. doi: 10.1097/01.mcg.0000155549.29643.ad. [DOI] [PubMed] [Google Scholar]
- 3.Alpini G, McGill JM, LaRusso NF. The pathobiology of biliary epithelia. Hepatology. 2002;35:1256–1268. doi: 10.1053/jhep.2002.33541. [DOI] [PubMed] [Google Scholar]
- 4.Dong H, Strome S, Salomao DR, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 5.Yamazaki T, Akiba H, Iwai H, Matsuda H, Aoki M, Tanno Y, Shin T, Tsuchiya H, Pardoll DM, Okumura K, et al. Expression of programmed death 1 ligands by murine T cells and APC. J. Immunol. 2002;169:5538–5545. doi: 10.4049/jimmunol.169.10.5538. [DOI] [PubMed] [Google Scholar]
- 6.Dong H, Chen XM. Immunoregulatory role of B7-H1 in chronicity of inflammatory responses. Cell. Mol. Immunol. 2006;3:179–187. [PMC free article] [PubMed] [Google Scholar]
- 7.Krambeck AE, Dong H, Thompson RH, Kuntz SM, Lohse CM, Leibovich BC, Blute ML, Sebo TJ, Cheville JC, Parker AS, Kwon ED. Survivin and B7-H1 are collaborative predictors of survival and represent potential therapeutic targets for patients with renal cell carcinoma. Clin. Cancer Res. 2007;13:1749–1756. doi: 10.1158/1078-0432.CCR-06-2129. [DOI] [PubMed] [Google Scholar]
- 8.Zhu B, Guleria I, Khosroshahi A, Chitnis T, Imitola J, Azuma M, Yagita H, Sayegh MH, Khoury SJ. Differential role of programmed deathligand 1 [corrected] and programmed death-ligand 2 [corrected] in regulating the susceptibility and chronic progression of experimental autoimmune encephalomyelitis. J. Immunol. 2006;176:3480–3489. doi: 10.4049/jimmunol.176.6.3480. [DOI] [PubMed] [Google Scholar]
- 9.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 10.Zang X, Allison JP. The B7 family and cancer therapy: costimulation and coinhibition. Clin. Cancer Res. 2007;13:5271–5279. doi: 10.1158/1078-0432.CCR-07-1030. [DOI] [PubMed] [Google Scholar]
- 11.Dong H, Zhu G, Tamada K, Flies DB, van Deursen JM, Chen L. B7-H1 determines accumulation and deletion of intrahepatic CD8+ T lymphocytes. Immunity. 2004;20:327–336. doi: 10.1016/s1074-7613(04)00050-0. [DOI] [PubMed] [Google Scholar]
- 12.Mataki N, Kikuchi K, Kawai T, Higashiyama M, Okada Y, Kurihara C, Hokari R, Kawaguchi A, Nagao S, Kondo T, et al. Expression of PD-1, PD-L1, and PD-L2 in the liver in autoimmune liver diseases. Am. J. Gastroenterol. 2007;102:302–312. doi: 10.1111/j.1572-0241.2006.00948.x. [DOI] [PubMed] [Google Scholar]
- 13.Kamihira T, Shimoda S, Nakamura M, Yokoyama T, Takii Y, Kawano A, Handa M, Ishibashi H, Gershwin ME, Harada M. Biliary epithelial cells regulate autoreactive T cells: implications for biliary-specific diseases. Hepatology. 2005;41:151–159. doi: 10.1002/hep.20494. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 16.Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol. Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Chen XM, Splinter PL, O’Hara SP, LaRusso NF. A cellular miRNA, let-7i, regulates toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J. Biol. Chem. 2007;282:28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grubman SA, Perrone RD, Lee DW, Murray SL, Rogers LC, Wolkoff LI, Mulberg AE, Cherington V, Jefferson DM. Regulation of intracellular pH by immortalized human intrahepatic biliary epithelial cell lines. Am. J. Physiol. 1994;266:G1060–G1070. doi: 10.1152/ajpgi.1994.266.6.G1060. [DOI] [PubMed] [Google Scholar]
- 20.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, Palfi A, Farrar GJ. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das S, Suarez G, Beswick EJ, Sierra JC, Graham DY, Reyes VE. Expression of B7-H1 on gastric epithelial cells: its potential role in regulating T cells during Helicobacter pylori infection. J. Immunol. 2006;176:3000–3009. doi: 10.4049/jimmunol.176.5.3000. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen XM, Gores GJ, Paya CV, LaRusso NF. Cryptosporidium parvum induces apoptosis in biliary epithelia by a Fas/Fas ligand-dependent mechanism. Am. J. Physiol. 1999;277:G599–G608. doi: 10.1152/ajpgi.1999.277.3.G599. [DOI] [PubMed] [Google Scholar]
- 26.Ding H, Wu X, Gao W. PD-L1 is expressed by human renal tubular epithelial cells and suppresses T cell cytokine synthesis. Clin. Immunol. 2005;115:184–191. doi: 10.1016/j.clim.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Filippatos G, Ang E, Gidea C, Dincer E, Wang R, Uhal BD. Fas induces apoptosis in human coronary artery endothelial cells in vitro. BMC Cell Biol. 2004;5:6. doi: 10.1186/1471-2121-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Boxel-Dezaire AH, Stark GR. Cell type-specific signaling in response to interferon-γ. Curr. Top. Microbiol. Immunol. 2007;316:119–154. doi: 10.1007/978-3-540-71329-6_7. [DOI] [PubMed] [Google Scholar]
- 29.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burgem CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 30.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darnell JE., Jr. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 32.Wesoly J, Szweykowska-Kulinska Z, Bluyssen HA. STAT activation and differential complex formation dictate selectivity of interferon responses. Acta Biochim. Pol. 2007;54:27–38. [PubMed] [Google Scholar]
- 33.Kaneko T, Tamai K, Matsuzaki Y, Yamazaki T, Nakano H, Kon A, Hashimoto I, Hanada K, Kaneda Y, Uitto J. Interferon-γ downregulates expression of the 230-kDa bullous pemphigoid antigen gene (BPAG1) in epidermal keratinocytes via novel chimeric sequences of ISRE and GAS. Exp. Dermatol. 2006;15:308–314. doi: 10.1111/j.0906-6705.2006.00408.x. [DOI] [PubMed] [Google Scholar]
- 34.Kraus J, Börner C, Lendeckel U, Höllt V. Interferon-γ down-regulates transcription of the μ-opioid receptor gene in neuronal and immune cells. J. Neuroimmunol. 2006;181:13–18. doi: 10.1016/j.jneuroim.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 35.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoenborn JR, Wilson CB. Regulation of interferon-γ during innate and adaptive immune responses. Adv. Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- 37.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Bezerra JA. Biliary atresia: translational research on key molecular processes regulating biliary injury and obstruction. Chang Gung Med. J. 2006;29:222–230. [PubMed] [Google Scholar]