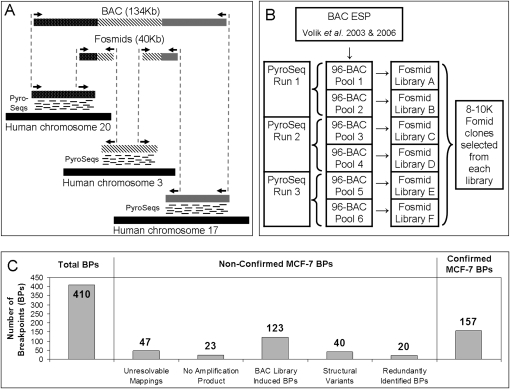
Figure 1.
(A) An illustration of the principle of the method. Breakpoints within a BAC containing segments from chromosomes 20, 3, and 17 are detected using a combination of “bridging” and “outlining” steps. The bridging step maps fosmid end-sequences onto the reference genome. The outlining step maps short tags (labeled “PyroSeqs”) using 454 technology from the BAC (in practice a pool of BACs) onto the reference genome. The results of bridging and outlining jointly allow precise mapping of breakpoints and reconstruction of rearranged BACs. (B) Organization of the mapping experiment. The nonredundant collection of 552 rearrangement containing BACs, 17 normal BAC negative controls, and seven positive controls was arrayed in six 96-well plates and pooled as indicated. Three 454 sequencing reactions (involving BACs pooled from plate pairs) produced tags for the purpose of outlining. Six fosmid libraries (one from each 96-well plate pool of BACs) were constructed for Sanger-based sequencing of fosmid ends and bridging. (C) Bar charts detailing the classification of detected MCF-7 breakpoints.