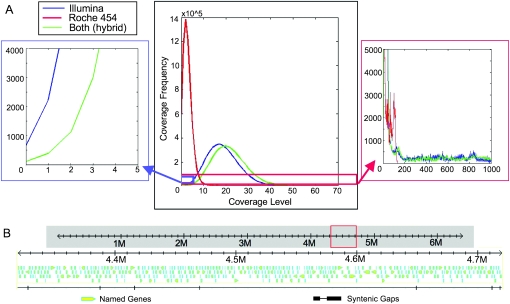
Figure 1.
Syntenic reassembly of PtoDC3000 demonstrates that few bases were not sequenced and that the PtoDC3000 genome has multiple stretches of repetitive sequence. (A) Histogram (center) showing the number of bases (×105) of the PtoDC3000 reference genome that are covered by sequence data at a given coverage level (x-axis). One lane of Illumina GA1 sequence, 1/4-plate of 454 sequence, and the combination of both sequence sets are shown. Only 656 bp remain unsequenced from the single lane of Illumina sequence (blue line), whereas 535,820 bp remain unsequenced from the plate of 454 long reads (red line). Combination of both sequence types (green) reduces this to 107 bp (blue box, center, expanded at left). PtoDC3000 has many repetitive regions over 35 bp in length (red box, center, expanded at right). This will cause difficulty in de novo assembly of only Illumina reads, as repeats will break assembly by most methods. However, the longer 454 reads and 454 paired-end reads partially ameliorate this problem. (B) Example of a typical 400-kbp genomic region with ORFs derived from the reference sequence as yellow arrows and unsequenced bases noted as ticks in the syntenic gaps line. The missing bases are scattered randomly and tend to be only one or a few bases in length.