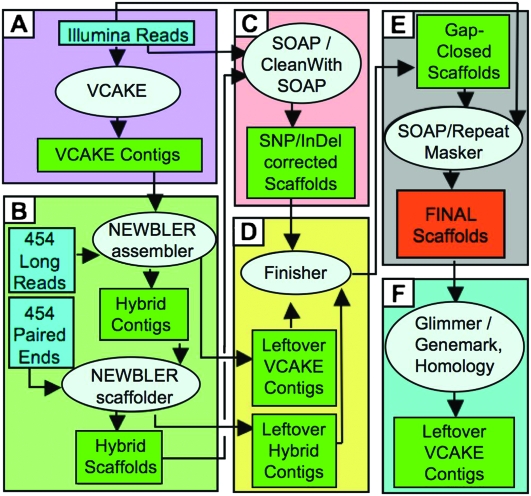
Figure 2.
Hybrid genome assembly pipeline. (A) Illumina reads are de novo assembled with VCAKE into VCAKE contigs. (B) Newbler then assembles VCAKE contigs and 454 long reads into hybrid contigs. The Newbler scaffolder orients the hybrid contigs into larger hybrid scaffolds using 454 paired-end data. (C) Hybrid scaffolds are cleaned of 1- to 2-bp indels using Illumina read depth; (D) longer gaps within scaffolds are filled with unused VCAKE and hybrid contigs in Finisher. (E) Polymorphism and coverage in the scaffolds are used to identify any putative repeat regions (see also Supplemental Figs. 2, 3). (F) Final scaffolds are used as templates for gene prediction using both homology and gene prediction software (Glimmer and GeneMark). See Methods for a complete description.