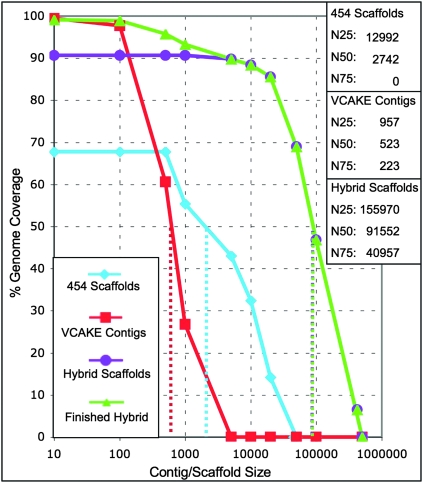
Figure 3.
Hybrid assembly outperforms assembly of either Illumina or 454 data sets alone. We reassembled the PtoDC3000 genome de novo to validate our assembly approach. For a given reassembly, the percentage of the reference PtoDC3000 genome (y-axis) covered by contigs of a given size (x-axis) is shown. The ∼3× coverage 454-only assembly (blue diamonds) reassembles only ∼70% of the PtoDC3000 genome, with most scaffolds in the 1- to 10-kb range. De novo assembly of the ∼26.3× Illumina read coverage only using the VCAKE assembler (red squares) leads to essentially complete genomic coverage, but in small contigs of less than 100 kbp. Hybrid assembly combining both technologies (purple circles) leads to 90% genomic coverage with hybrid contigs over 500 kbp. The subsequent finishing step incorporating unscaffolded contigs (Supplemental Fig. 1) leads to final coverage of 99.1% of the PtoDC3000 reference genome (green triangles). N50 values (dotted lines) increase at each step.