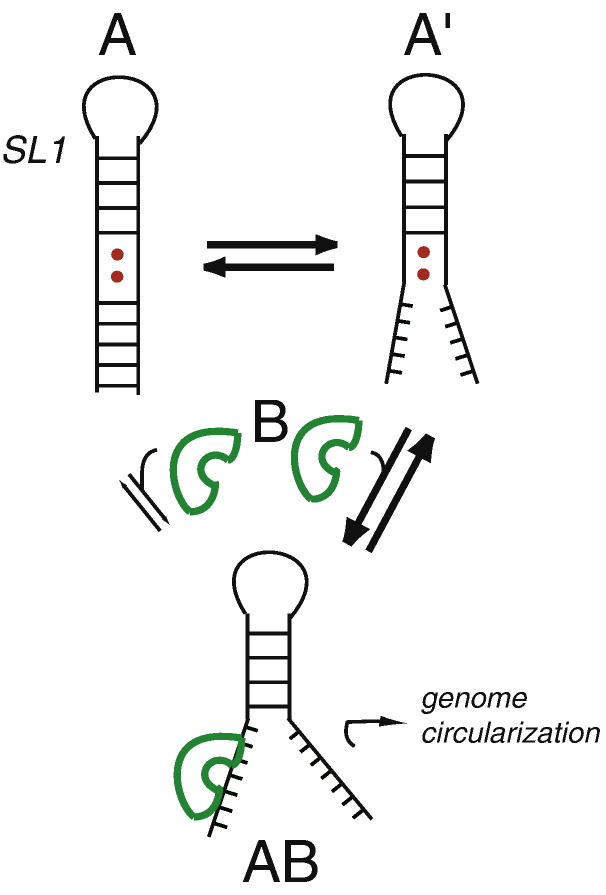
Fig. 9.
Model of a dynamic SL1 that is consistent with the functional and structural data presented here. The fully based-paired SL1 (A; modeled by the ΔA35 RNA) exists in equilibrium with one or more higher-energy conformers (A′; WT* and ΔA35 second-site revertants) that are partially unfolded or that experience dynamic destabilization as a result of noncanonical pairing. A hypothetical protein (B) binds to both A and A′ to form the same partially unwound AB complex, but the affinity of B for A′ will be higher, since the full energetic cost of unfolding the lower stem will not have to be paid; this interaction then mediates a long-distance RNA–RNA, RNA–protein, or protein–protein interaction, which is crucial for the viral replication.