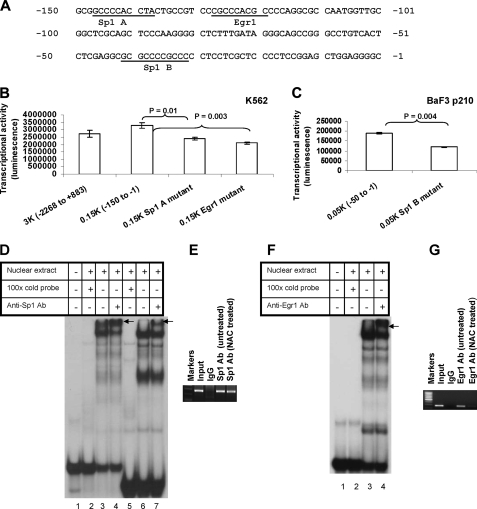
FIGURE 3.
Analysis of Sp1 and Egr1 binding sites in the Fyn promoter and their ROS dependence. A, the nucleotide sequence of the human Fyn promoter region (-150 to -1) that has high basal transcription activity. Numbering is relative to the transcription start site (+1). Putative binding sites for transcription factors Sp1 and Egr1 are indicated on the sequences. B, mutational analysis of the putative Sp1 A-binding site and Egr1-binding site of the Fyn promoter. Individual mutagenesis that destroyed Sp1 A and Egr1 putative binding sites were introduced to the pFynPromoterCheck plasmid by method described under “Experimental Procedures.” The Egr1 binding site was deleted, whereas the Sp1 binding site was mutated as described under “Experimental Procedures.” The mutant constructs were transfected into K562 cells. Renilla luciferase activity was normalized to firefly luciferase activity. C, mutational analysis of the putative Sp1 B-binding site of Fyn promoter in BaF p210 cells. Site-directed mutagenesis destroyed the Sp1 B putative binding site in the -50 to -1 region of the Fyn promoter construct. Following transfection of the mutant sequence into BaF3 p210 cells, dual-luciferase assays were used to measure promoter activity. D, gel shift assay reveals K562 cell nuclear proteins bind to the probe containing a putative Sp1 binding site. The 32P-labeled oligonucleotide probe (-150 to -121 bp) was incubated alone, with nuclear extract, with nuclear extract in the presence of excess unlabeled oligonucleotide, or with nuclear extract and a Sp1 antibody. Arrows indicate the supershifted complexes. Lanes 1-4 contain a 30-bp probe of the Fyn promoter, while lanes 5-7 contain a 22-bp Sp1 consensus probe (from Promega). E, chromatin immunoprecipitation of Sp1 with the Fyn promoter in K562 cells. Bands indicate PCR products targeting the -250 to -1 region of the promoter. 5 μg of anti-Sp1 or rabbit IgG antibody or 1/100 of input was separately added in the assay. F, gel shift assay of K562 cell nuclear proteins bind to a probe containing a putative Egr1 binding site. The 32P-labeled oligonucleotide probe (-140 to -111 bp) was incubated alone, with nuclear extract, with nuclear extract in the presence of excess unlabeled oligonucleotide, or with nuclear extraction and anti-Egr1 antibody. Arrows indicate the supershifted complexes. G, chromatin immunoprecipitation of Egr1 with the human Fyn promoter in K562 cells. Bands indicate PCR products targeting the -250 to -1 region of the Fyn promoter. K562 cells were untreated or treated with 24 mm NAC for 24 h. 5 μg of anti-Egr1 or rabbit IgG antibody or 1/100 of input was separately added in the assay. Every result shown is representative of three independent experiments.