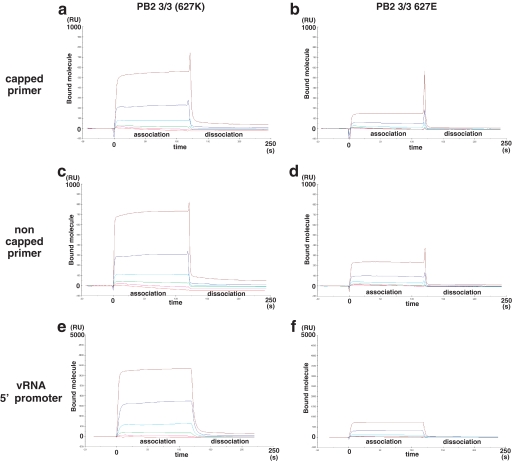
FIGURE 3.
The role of lysine 627 in the binding of PB2 3/3 to RNA oligomers. RNA binding analyses of PB2 3/3 and PB2 3/3 Glu627 proteins were conducted using a surface plasmon resonance assay. Various concentrations (0.015, 0.044, 0.13, 0.40, 1.2, and 3.6 μm) of recombinant PB2 3/3 and PB2 3/3 Glu627 proteins were incubated with the RNA oligonucleotides that had been immobilized on a sensor chip. The 5′-vRNA promoter (5′-AGUAGAAACAAGGCC-3′), a capped primer (5′-m7GmAAAUACUCAAG-3′), or a non-capped primer (5′-AmAAUACUCAAG-3′) was tested for PB2 3/3 domain protein binding. The vertical axis in each panel indicates the resonance units (RU), which reflect the number of attached molecules observed in real time (horizontal axis, full scale is 250 s). The red, magenta, green, cyan, blue, and brown lines indicate the binding curves for the 0.015, 0.044, 0.15, 0.44, 1.2, and 3.6 μm protein concentrations, respectively. The data shown indicate the resonance units after subtraction of the background values (no RNA measurements). The labels association and dissociation represent the buffer in the presence or absence of the input molecules, respectively. Affinity analysis of the binding of oligomer RNA to recombinant PB2 3/3 protein was performed, and the calculated Kd and Rmax values are shown in supplemental Table 1.