Abstract
The PrP gene encodes the cellular isoform of the prion protein (PrPc) which has been shown to be crucial to the development of transmissible spongiform encephalopathies (TSEs). PrP knock-out mice, which do not express endogenous PrPc, exhibit resistance to TSE disease. The regulation of PrP gene expression represents, therefore, a crucial factor in the development of TSEs. Two sequence motifs in the PrP promoter (positions –287 to –263 from transcriptional start) were previously reported as being highly conserved, and it was suggested that they represent binding sites for as yet unidentified transcription factors. To test this hypothesis, binding of nuclear proteins was analyzed by electrophoretic mobility shift assays using ovine or murine cells and tissues with radiolabeled DNA probes containing the conserved motif sequences. Specific binding was observed to both motifs, and polymorphic variants of these motifs exhibited differential binding. Two proteins bound to these motifs were identified as the Yin Yang 1 (YY1) (motif 1) and E4BP4 (motif 2) transcription factors. Functional promoter analysis of four different promoter variants revealed that motif 1 (YY1) was associated with inhibitory activity in the context of the PrP promoter, whereas motif 2 (E4BP4) was linked to a slight enhancing activity. This represents the first demonstration of binding of nuclear factors to two highly conserved DNA sequence motifs within mammalian PrP promoters. The action of these factors on the PrP promoter is haplotype-specific, leading us to propose that the prion protein expression pattern and, with it, the distribution of TSE infectivity may be associated with PrP promoter genotype.
The ovine PrP gene (PRNP in human, prn-p in mice) encodes the prion protein (PrPC),4 which is a crucial component of prion diseases, also known as transmissible spongiform encephalopathies (TSEs). This group of diseases include scrapie in sheep, bovine spongiform encephalopathy in cattle, and Creutzfeldt-Jakob disease in humans (1). The hallmark of TSEs is the aggregation of a pathological isoform PrPSc of the cellular PrPC protein (2). PrPC is highly expressed in neurons of the brain and also found in many other tissues, especially the lymphoreticular system. Prn-p knock-out mice, which do not express endogenous PrPC, exhibit resistance to TSE disease (3). Transgenic mice with differing copy numbers of the prn-p gene show a clear relationship between expression level and disease progression, characterized by changes in incubation periods (4). It is assumed that the TSE agent can only replicate in cells expressing PrPC, so that up- or down-regulation of the PrP promoter can have consequences for the distribution of the agent in the host and, therefore, the risk of transmission; for example, from consumption of specific animal parts or blood transfusion in humans (5).
Beside its key role in disease, the physiological function of PrPC remains elusive with signal transduction, synaptic transmission, neuroprotection, and immunoregulation among the proposed properties (6). The subtle phenotypes observed in PrPC knock-out mice are as diverse as disturbance of circadian rhythm (7) and abnormalities in tooth development (8). A detailed understanding of the regulation of the ovine PrP gene expression will help to clarify its physiological and disease-related physiological role.
Surprisingly, given its importance in the development of TSEs, very little is known about the DNA motifs involved in the regulation of the PrP gene. Regulatory sequences have been mapped to a region ∼90 base pairs upstream of the rat and bovine genes that appears to depend for activity on elements in intron 1 (9, 10) and exon 1 (11). This upstream region is rich in consensus binding sites for SP1 that are highly conserved between species (12). Two polymorphisms in the bovine PrP gene promoter have been shown to alter the binding of transcription factors RP58 and SP1, thereby changing the expression of reporter genes (13). There are also four highly conserved sequences, designated motifs 1–4, located within the region –220 to –290 (14). Of particular interest is the presence of a T to C polymorphism within motif 1 (–278 to –290) of the ovine PrP promoter (15, 16). Additionally, motif 2 (–266 to –278) contains a species-specific sequence variation where the ruminant PrP promoter contains a C (TTACGTAA), whereas the non-ruminant promoter containsaTatthe same position (TTATGTAA).
The aim of this study was to identify factors that bind to the ovine PrP gene promoter and to determine their role in transcriptional regulation. Our data reveal binding of the transcription factors YY1 to motif 1 and of E4BP4 to motif 2. Whereas YY1 acted as an inhibitor but only bound to one PrP haplotype, E4BP4 acted as an activator of transcription binding with different affinities to all PrP haplotypes.
EXPERIMENTAL PROCEDURES
DNA Probes—Complimentary pairs of oligonucleotides (Eurofins MWG, Germany) containing the conserved motif sequences were annealed forming short double-stranded DNA fragments (probes) that were radiolabeled with [γ-32P]ATP (Amersham Biosciences) using T4 polynucleotide kinase (Promega, Southampton, UK). The following probes (only top strand sequence given) were used: M1T, 5′-TAATCTTTCATTTTCTCCAT-3′; M1C, 5′-TAATCTTCCATTTTCTCCAT-3′; M2C, 5′-TTCTCCATTACGTAACGAGA-3′; M2T, 5′-TTCTCCATTATGTAACGAGA-3′; M1CM2C, 5′-TAATCTTCCATTTTCTCCATTACGTAACGAGA-3′; NC-1, 5′-ATTCGATCGGGGCGGGGCGAGC-3′.
Nuclear Extracts and Electrophoretic Mobility Shift Assay (EMSA)—DNA probes were incubated in binding assays with nuclear extracts (NEs) prepared from ovine cell cultures (cerebellar-derived cell culture IS120Cer, liver-derived cell culture IS120Liv, brain cell cultures sA80BR and pA80BR), murine neuroblastoma cell line N2a, human cervical cancer HeLa cells (Promega), or from tissues derived from C57BL mice and from sheep using the NE-PER nuclear extraction reagent (Pierce). EMSAs were performed using the gel shift assay kit (Promega). Reactions consisted of 2 μl of 5× binding buffer (250 mm NaCl, 50 mm Tris-HCl, pH 7.5, 2.5 mm EDTA, 2.5 mm dithiothreitol, 5 mm MgCl2, 20% glycerol, 0.5 μg poly(dI·dC)·poly(dI·dC)), 1–2 μl (10–20 μg) NE, and dH2O to a total volume of 10 μl incubated at 25 °C for 10 min. To the reaction mixture 1 μl of 32P labeled double-stranded DNA probe (0.035 pmol, ∼10,000–50,000 cpm/10–100 fmol) was added and further incubated at 25 °C for 20 min. For competition experiments a 100-fold molar excess of unlabeled competitor and/or non-competitor (NC-1) double-stranded DNA probes were added. The reactions were electrophoresed in a 4% non-denaturing polyacrylamide gel in 0.5× Tris-buffered EDTA at 250 V at room temperature for 1 h, dried, and subjected to autoradiography.
Antibody Competition Assay—EMSAs were performed as before except for the addition of the antibody (1–2 μg, Transcruz™ Gel Supershift antibody; Santa Cruz Biotechnology, CA) immediately after the appropriate 32P-labeled probe was added to the reaction mixture. The reaction was incubated at room temperature for 45 min before gel electrophoresis. The antibodies used were the rabbit polyclonal antibody H-414 (anti-YY1, specificity demonstrated for YY1 of mouse, rat, and human origin) and the goat polyclonal antibody V-19 (anti-E4BP4, specificity demonstrated for E4BP4 of mouse, rat, and human origin).
Transcription Factor Analysis Software—Transcription factor binding sites were analyzed with the TFSEARCH program (Version 1.3) using the TRANSFAC matrix Table 2.5 (19).
Promoter Constructs—The plasmid pNPU10 as described in O'Neill et al. (15) contains a sheep promoter fragment equivalent to positions 5226–5747 of the PrP sequence (GenBank™ accession number U67922). pNPU10 has the combination of C5382 in motif 1 and G5622 (SP1 site) as described in O'Neill et al. (16). In vitro mutagenesis with the QuikChange® site-directed mutagenesis kit and XL1-Blue Supercompetent cells (Stratagene) was used to change C5382 to T5382 on pNPU10 to create plasmid pNPU6TC. This plasmid was used in another round of in vitro mutagenesis to change CG into AA at positions 5397/5398 to create plasmid pNPU2TAA. The promoter fragment was PCR-amplified from all three plasmids with oligonucleotides ProMluWg1 (5′-TCAGACGCGTTGACGGCAGGTGATGGCTAA-3′), ProBglWg2 (5′-AGTTAGATCTGCGGCTGTCAGCGACT-3′), and PCR products were purified with QIAquick Gel extraction kit (Qiagen) and subcloned into pGem-T-Easy. pGemT-2TAA was then changed by in vitro mutagenesis to C in position 5382 to create pGEMT-7CAA. The four different promoter fragments were excised with restriction enzymes, MluI and BglII (New England Biolabs) and inserted into MluI and BglII sites in plasmid pGL3Basic (Promega). The promoter sequences of the four resulting plasmids (p110-1, p7CAA, p6TC, p2TAA) were verified by sequencing of both strands.
Cell Culture—Ovine cell cultures sA80BR and pA80BR, derived from fetal sheep brain tissue, were grown as described (17); they contained a mixture of neuronal and glial cells. Two primary ovine cell cultures derived from the cerebellum (IS120Cer) and liver (IS120Liv) were produced from a 1.5-year-old Icelandic short-tailed sheep (Ovis brachyuran borealis pall) by standard methods (18). In short, tissue was recovered in ∼2-cm2 segments from brain and liver and mechanically macerated into smaller pieces. After incubation with 0.25% trypsin at 37 °C for 30 min, the cell debris was pelletted, and the suspended cells (lysate) were removed. The cell pellet was resuspended in fresh trypsin solution, and the process was repeated three times. All cells (lysates) were resuspended in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal calf serum (Invitrogen). Aliquots of the lysates in Dulbecco's modified Eagle's medium were added to pre-gelatinized (0.1%) 25-cm2 flasks and incubated at 37 °C, 5% CO2. Growing cell colonies were occasionally dislodged to reach confluent growth before passage. Murine neuroblastoma cells (N2a, ATTC number CCL131) were provided by Dr. Herbert Baybutt, Neuropathogenesis Division, Roslin Institute. N2a cells were maintained in monolayer culture in Dulbecco's modified Eagle's minimum essential medium glucose (1 g/liter), 2 mm l-glutamine, 1 mm sodium pyruvate (Fisher) with the addition of 10% (v/v) fetal bovine serum (Fisher). N2a cells were maintained at 37 °C, and sA80Br and pA80BR cells were maintained at 33 °C; all cells were grown without antibiotics in a humidified atmosphere of 5% CO2.
Transient Transfections—N2a cells on 6-well plates at ∼90% confluence were transiently transfected with plasmids using Lipofectamine 2000 reagent (Invitrogen) in a 500-μl transfection mixture containing 250 ng of plasmid DNA. Cells were incubated for 4 h in the transfection mixture, which was then replaced with 2 ml of normal growth medium. Cells were analyzed 48 h post-transfection. Three transfections per construct series were performed. Results were averaged, and Student's t test was applied for statistical analysis.
Luciferase Assays—N2a cells were washed once with phosphate-buffered saline and lysed with 500 μl of Passive Lysis Buffer (Promega). Cell lysis was carried out for 15 min with shaking, and cell debris was removed by centrifugation at 13,000 × g for 5 min at 4 °C. Firefly and Renilla luciferase activities were determined in extracts of transfected cells using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol.
RESULTS
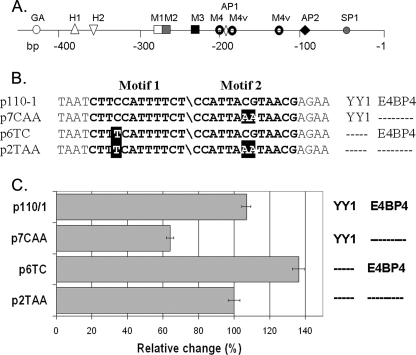
Motif 1 and motif 2 are highly conserved sequences (Fig. 1) located at ∼280–300 base pairs upstream of the ovine PrP promoter transcription start site (see Fig. 5A).
FIGURE 1.
Transcription factor binding sites and conserved motifs in PrP promoter. Alignment of motifs 1 and 2 (in bold) for nine species. Sheep T, T-haplotype of PrP gene promoter; Sheep C, C-haplotype of PrP gene promoter. The dashes indicate alignment gap.
FIGURE 5.
Luciferase reporter gene assays. A, diagram of ovine core promoter as used in transfection experiments. GA, putative GATA-1 factor binding site; H1 and H2, heat shock element motifs; M1, M2, M3, and M4, conserved motifs as defined in Westaway et al., (14); M4v, variants of M4 motifs; AP1, AP2, and SP1, putative binding sites for transcription factors AP1, AP2, and SP1. B, four plasmid constructs containing the PrP gene promoter intact or with mutations in either motif 1 (YY1 binding site) or motif 2 (E4BP4 binding site) in front of the luciferase open reading frame were used to transfect N2a neuroblastoma cells. C, mean relative luciferase activity and S.D. from three experiments are shown. Columns on the right site of the diagram indicate the presence or absence of the binding sequences for YY1 and E4BP4, respectively.
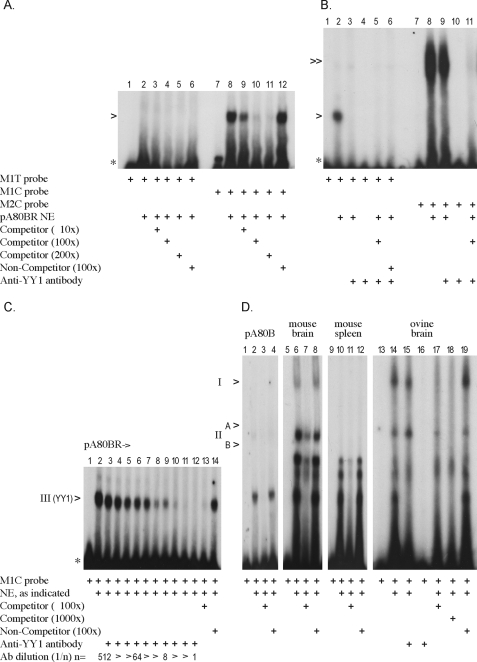
Motif 1—EMSAs were performed comparing the motif 1 core sequence CTTTCATTTTCT (M1T probe) and the naturally occurring variant motif 1 sequence CTTCCATTTTCT (M1C probe) for their ability to form DNA-protein complexes when incubated with ovine sA80BR cell culture NE. Only the M1C probe binds with high affinity to a nuclear protein shown by the shift of the probe to a higher molecular weight (lane 8, Fig. 2A). This binding was competed out by the addition of increasing concentrations (10-, 100-, and 200-fold molar excess) of unlabeled M1C probe (competitor) (lanes 9–11, Fig. 2A). Competition with the unlabeled NC-1 probe (non-competitor) had no effect on the formation of the M1C DNA-protein complex, verifying that binding to the variant motif 1 site was sequence-specific (lane 12, Fig. 2A). In contrast, the M1T probe failed to show a band shift with the ovine sA80BR cell culture NE (lane 2, Fig. 2A), although a much weaker signal similar to the M1C probe was occasionally detectable with ovine nuclear extract. This differential binding was also observed with both the M1C and the M1T probes when this assay was performed using the pA80BR, IS120Liv, or IS120Cer NEs (data not shown).
FIGURE 2.
Motif 1 EMSAs. A, motif 1 (M1C and M1T) probes incubated with pA80BR NE. The EMSA was performed twice per sheep cell extract (pA80BR, sA80BR, IS120Cer, IS120Liv); only data for pA80BR are shown. The intensity of the M1C-protein complex was consistently as high as shown, whereas the M1T-protein complex varied between 0 and 10% that of the M1C complex (indicated with >). B, competition EMSA; Lanes 1–6, variant motif 1 probe (M1C) incubated with pA80BR NE in the presence of anti-YY1 antibody (Ab) and unlabeled probes as indicated; lanes 7–11, control experiment: motif 2 probe (M2C) incubated with pA80BR NE in the presence of anti-YY1 antibody (Motif 2 specific complex indicated with ≫). The antibody competition assay was performed a total of 4 times in different combinations as shown in B–D. C, competition EMSA. Variant motif 1 probe (M1C) incubated with pA80BR NE in the presence of increasing concentrations of anti-YY1 antibody. D, tissue extracts (performed twice with comparable results). Motif 1 probe (M1C) incubated with ovine pA80BR NE as control, ovine and murine brain tissue-derived Nes, and murine spleen tissue-derived NE. Specific complexes I (Fy-I), IIA and IIB (Fy-II), and III (YY1) were detected in tissues. The anti-YY1 antibody was added immediately after the addition of 32P probe and incubated at room temperature for 45 min. Competitor, unlabeled probe M1C; Non-competitor, unlabeled probe NC-1. 10×, 100×, 200×, or 1000× indicates the addition of a 10, 100, 200, or 1000-fold molar excess of unlabeled probe. *, unbound 32P-labeled probe.
A search of transcription factor binding site databases revealed the
highest similarity between the consensus motif of transcription factor Yin
Yang 1 (YY1) and the variant motif 1 sequence
(20). To determine whether YY1
was actually part of the motif 1 DNA-protein complex, we employed an anti-YY1
antibody in competition assays performed with the M1C probe incubated with
pA80BR NE. The addition of the anti-YY1 antibody resulted in the abolition of
the initial M1C DNA-protein complex (lane 3,
Fig. 2B and compare
with lane 2, Fig.
2B). The competition assay was repeated with murine N2a
cell NE with the same result (data not shown). The M1C probe was then
incubated with ovine pA80BR NE, and increasing concentrations of the anti-YY1
antibody were added (Fig.
2C). The amount of the DNA-protein complex (lane
2, Fig. 2C)
gradually decreased as more anti-YY1 antibody was added until it was no longer
detectable at a  dilution of
the antibody (lanes 3–12,
Fig. 2C). This
concentration-dependent reduction provided further evidence that it was indeed
the transcription factor YY1 that was bound to the variant motif 1 site. The
competition assay was repeated with the M1C probe and an unrelated anti-E4BP4
antibody with no apparent effect on the M1C DNA-protein complex, indicating
that the reaction seen with the anti-YY1 antibody was likely to have been
caused by the antibody itself and not a component of the antiserum.
Specificity for the M1C probe was evident from the fact that no change in the
M2C DNA-protein complex was observed when the ruminant motif 2 probe M2C was
incubated with the anti-YY1 antibody (lane 9,
Fig. 2B).
dilution of
the antibody (lanes 3–12,
Fig. 2C). This
concentration-dependent reduction provided further evidence that it was indeed
the transcription factor YY1 that was bound to the variant motif 1 site. The
competition assay was repeated with the M1C probe and an unrelated anti-E4BP4
antibody with no apparent effect on the M1C DNA-protein complex, indicating
that the reaction seen with the anti-YY1 antibody was likely to have been
caused by the antibody itself and not a component of the antiserum.
Specificity for the M1C probe was evident from the fact that no change in the
M2C DNA-protein complex was observed when the ruminant motif 2 probe M2C was
incubated with the anti-YY1 antibody (lane 9,
Fig. 2B).
Binding to the motif 1 probes was also analyzed with NEs prepared from ovine and murine brain tissue, and three specific M1C DNA-protein complexes (I, II, and III, lanes 6 and 10, Fig. 2D) were observed, the smallest of which (complex III) appeared to be similar to the YY1 complex observed with the ovine pA80BR cell culture NE (lane 2, Fig. 2D). The assay was repeated using the anti-YY1 antibody as a blocking agent as described above (lanes 9–15, Fig. 2D). Only complex III was blocked (lane 15, Fig. 2D), confirming that the YY1 transcription factor also binds to the M1C probe in the ovine brain NE. A similar blocking reaction on other promoters has been reported with the same anti-YY1 antibody (21), although a definite supershift reaction has also been noted (22). The two additional complexes were not affected by the YY1 antibody added either before or after the addition of the DNA probe, evidently representing the binding of other as yet unidentified proteins. These complexes are not related to each other, as complex I is absent with the M1T probe, whereas complex II binds to both the M1C and M1T probes equally (data not shown). In addition, complex I appears to be absent in nuclear extracts prepared from murine spleen (Fig. 2D, lanes 9–12) and liver tissue (data not shown), indicating that this factor may be brain-specific.
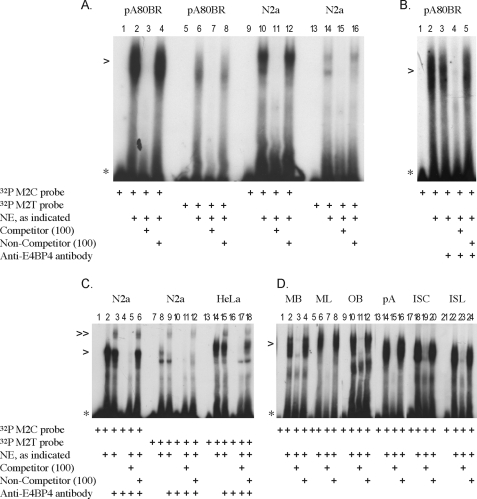
Motif 2—The ruminant (sheep, goat, cattle, deer) motif 2 core sequence (TTACGTAA) differs from the non-ruminant (mouse, Syrian hamster, human, and rat) motif 2 sequence by a single base change of C to T (TTATGTAA) (Fig. 1). EMSAs showed that the ruminant motif 2 (M2C) probe was bound with high affinity by protein(s) present in the ovine pA80BR cell culture NE as well as in the murine N2a cell culture NE (lanes 2 and 10, respectively, Fig. 3A). In contrast, the non-ruminant motif 2 (M2T) probe was bound with much less affinity by what may be the same protein(s) based on comparisons of their electrophoretic mobilities (lanes 6 and 14, respectively, Fig. 3A). Binding was competed out by the addition of a 100-fold molar excess of unlabeled competitor probe (lanes 3, 7, 11, and 15, Fig. 3A). In contrast, binding was not affected by the addition of a 100-fold molar excess of unlabeled NC-1 probe (non-competitor) (lanes 4, 8, 12, and 16, Fig. 3A), indicating that the complexes are sequence-specific. The same results were observed with both the M2C and M2T probes when this assay was performed using the ovine sA80BR, IS120Liv, and IS120Cer NEs (data not shown).
FIGURE 3.
Motif 2 EMSAs. Each EMSA was performed at least twice per extract (pA80BR, sA80BR, N2a, HeLa, tissue extracts). A, ruminant motif 2 (M2C) and non-ruminant motif 2 (M2T) probes incubated with pA80BR and N2a NEs (specific complex indicated with >). B, competition EMSA; ruminant motif 2 (M2C) probe incubated with pA80BR NE in the presence of anti-E4BP4 antibody. C, competition EMSA; motif 2 (M2C and M2T) probes incubated with N2a and HeLa NEs in the presence of anti-E4BP4 antibody (supershifted Motif 2-E4BP4 antibody complex indicated with ≫). D, comparison of ruminant motif 2 (M2C) probe with ovine and murine tissue-derived NEs and ovine cell NEs. Nes. MB, mouse brain; ML, mouse liver; OB, ovine brain; pA, ovine pA80BR cells; ISC, ovine IS120Cer cells; ISL, ovine ISL120Liv. Anti-E4BP4 antibody was added immediately after the addition of 32P probe and incubated at room temperature for 45 min. Competitor, unlabeled probe M2C; non-competitor, unlabeled probe NC-1. 100× indicates the addition of a 100-fold molar excess of unlabeled probe.
A strong candidate for motif 2 binding was the E4BP4 transcription factor, which recognizes the palindromic motif of TTACGTAA (23). To determine whether the protein bound to motif 2 was E4BP4 we employed an anti-E4BP4 antibody in a supershift assay with ovine pA80BR nuclear extract. The supershift assays were carried out by adding the antibody either before (data not shown) or immediately after the addition of the radiolabeled probe (lanes 1–5, Fig. 3B). In addition, two different sets of incubation conditions were used, either for 45 min at room temperature (lanes 1–5, Fig. 3B) or overnight at +4 °C (data not shown). No supershift of the original M2C DNA-protein complex was observed with ovine pA80BR NE under any of the conditions tested (lanes 1–5, Fig. 3B). To verify that the conditions for a supershift were correct, the anti-E4BP4 antibody was incubated with murine N2a cell NE in combination with the M2C probe (Fig. 3C). In contrast to the ovine pA80BR cell NE experiments, a supershift occurred with the murine N2a cell NE (lane 3, Fig. 3C), and no supershift reaction was observed when the NE was omitted (lane 4, Fig. 3C). Furthermore, the initial shift and the supershift were both competed out by the addition of unlabeled M2C probe (competitor) but not with unlabeled NC-1 probe (non-competitor) (lanes 5 and 6, respectively, Fig. 3C). A similar supershift was observed for the M2T probe with the murine N2a cell NE, indicating that the E4BP4 transcription factor is able to bind to both the ruminant and non-ruminant forms of motif 2 in mouse cell NE (lane 9, Fig. 3C). In addition, a supershift was seen for the M2T probe with the human HeLa NE (lane 15, Fig. 3C). A complex similar in electrophoretic mobility to E4BP4 was observed with NEs derived from ovine and murine brain tissues and the M2C probe (lanes 2 and 10, respectively, Fig. 3D). A slight difference in electrophoretic mobilities was observed between the complexes from the murine brain and liver NEs (lanes 2 and 6, respectively, Fig. 3D) and also between the IS120Cer and IS120Liv NEs (lanes 18 and 22, respectively, Fig. 3D). The significance of these differences remains to be established; however, they may represent the binding of tissue-specific homo- and heterodimers to the same motif.
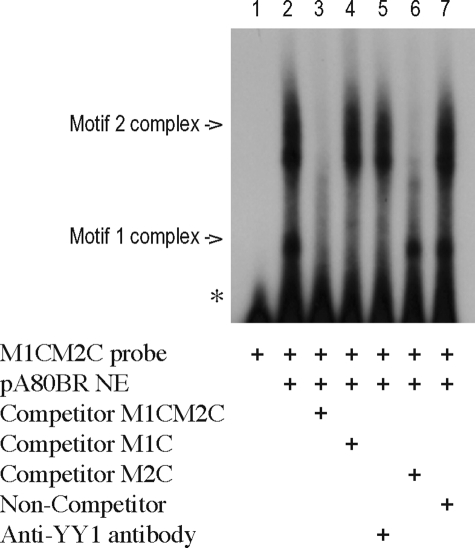
Analysis of Potential Interactions between Motif 1 and Motif 2 Binding Factors—In the ovine PrP promoter, motifs 1 and 2 are contiguous, and their proximity may influence the pattern of transcription factor binding. The ability of both factors to bind at the same time was investigated using a series of double-motif probes that contained the different combinations of motifs 1 and 2 and their variants. Based on the results from the single motif probe experiments, the strongest binding for both factors is predicted for the combination of the C-haplotype motif 1 and the ruminant motif 2 (M1CM2C probe). Two individual complexes were observed with the M1CM2C probe and pA80BR NE, which were consistent with the separate formation of a M1C or a M2C DNA-protein complex (lane 2, Fig. 4). Both complexes were competed out by the addition of a 100-fold molar excess of unlabeled M1CM2C competitor probe (lane 3, Fig. 4) but were not affected by the addition of a 100-fold molar excess of unlabeled NC-1 non-competitor probe (lane 7, Fig. 4). Competition with either one of the single motif probes M1C or M2C at a 100-fold molar excess led as expected to the inhibition of the formation of the motif 1 or motif 2 complexes, respectively, but not of both together (lanes 4 and 6, respectively, Fig. 4). In addition, only the smaller complex was specifically blocked after the addition of anti-YY1 antibody, demonstrating that this complex represented the binding of the YY1 transcription factor (lane 5, Fig. 4). At no time was an additional shift complex seen (indicative of the binding of both complexes to the promoter sequence at the same time), suggesting that the binding of these factors is mutually exclusive, most likely caused by steric hindrance with this close sequence motif arrangement.
FIGURE 4.
Double-motif probe EMSAs. Motif 1 and motif 2 combined probes (double-motif probe M1CM2C) with pA80BR NE. Anti-YY1 antibody was added immediately after the addition of 32P probe and incubated at room temperature for 45 min. Competitor, 100-fold molar excess of unlabeled probes as indicated; non-competitor, 100-fold molar excess unlabeled probe NC-1.
Promoter-Reporter Gene Assays—To verify that both motifs can modulate transcription, we generated four constructs in which the luciferase open reading frame was under the control of the PrP promoter. As shown in Fig. 5, ovine motif 2 was present or absent in combination with T-haplotype or C-haplotype motif 1 both in the arrangement and distance to each other as found normally in the ovine PrP promoter. N2a cells were transfected, and luciferase activity was measured for each construct (Fig. 5C). The activity for p2TAA (T-haplotype motif 1 without motif 2) was set to 100%. A relative reduction was observed in two of the other three combinations of motifs 1 and 2. A highly significant reduction to 64% (S.D. ± 6) (p = 0.001) was seen with construct p7CAA (C-haplotype motif 1 without motif 2), indicating that factor YY1 repressed transcription. In contrast, a significant increase to 136% (S.D. ± 10) was seen for construct p6TC (T-haplotype motif 1 with motif 2) which appears to reveal the activator role of, probably, E4BP4. There was no significant difference between p110/1 (C-haplotype motif 1 with motif 2) and p2TAA, which can be interpreted as an indication of interference of the transcription factors binding to motif 1 and 2.
DISCUSSION
The high conservation of short sequence motifs in the ruminant, rodent, and human PrP gene promoters was first described by Westaway et al. (14) but has never been investigated properly. Here we tested the hypothesis that motif 1 and motif 2 are transcription factor binding sites. Special emphasis was given to the fact that the affinity for these potential factors may be associated with ovine C- and T-haplotypes of motif 1 (15) and a species-specific sequence variation in motif 2.
The YY1 Transcription Factor Binds to the Variant Motif 1 Site—Our experiments with nuclear extract from cell cultures have shown that the motif 1 sequence (CTTTCATTTTCT), as published for human PRNP, mouse prn-p, and sheep PrP promoters, does not form a stable protein-DNA complex under the conditions used in the mobility shift assays. However, a single base pair change from T to C in motif 1, which has been found as a polymorphism in sheep (15, 16), appears to have a dramatic effect on protein binding. The sequence CTTCCATTTTCT shows strong, consistent, and highly specific binding with nuclear extracts derived from ovine and murine cell cultures. Furthermore, protein binding to motif 1 was confirmed by DNase I footprinting of a 526-bp ovine PrP gene promoter fragment from p110-1 using pA80BR extract.5 The competition assay performed with the anti-YY1 antibody led to specific blocking of the formation of the M1C DNA-protein complex, which leaves us to believe that the factor bound to the motif 1 C haplotype is the transcription factor YY1. YY1 is a zinc finger transcription factor with a predicted molecular mass of 44 kDa; its binding sequence is located on a number of mammalian gene promoters, in which it either acts as an activator of transcription (e.g. p53 gene) or a repressor (e.g. serum amyloid A1 gene) (20). The binding motif for YY1 contains an (A/C)CATNTT core sequence, so that the T-haplotype motif 1 sequence (TCATTTT) of the PrP gene does not fit the YY1 consensus binding site, whereas the C-haplotype motif 1 sequence (CCATTTT) forms a consensus YY1 binding motif (24, 25). Interestingly, a similar situation has been described within the human cystic fibrosis transmembrane conductance receptor gene, where a polymorphism CCA(A/T)ATT produces a new YY1 binding site within this gene promoter. The presence of this polymorphism causes a significant increase in the expression of the cystic fibrosis transmembrane conductance receptor protein, and the authors hypothesize that this may have a beneficial effect on patients with cystic fibrosis (26). In the ovine PrP promoter the binding of YY1 has, according to our in vitro assays, repressor activity, and this may lead in vivo to reduced expression of PrP. YY1 is able to control gene transcription by associating with other regulatory proteins, such as specificity protein-1 (SP1), present on distant areas of DNA, and it is thought that YY1 achieves this by bending the DNA backbone (27). The ovine PrP gene promoter contains a specificity protein-1 binding site (–50) in proximity to the YY1 motif (–290) only on the C-haplotype (all constructs tested here contain this SP1 site) whether a haplotype with YY1 but without the SP1 site exists and shows different expression regulation remains to be established.
Intriguingly, YY1 sites are commonly found close to or overlapping other binding sites, and repression of transcription is then achieved indirectly by displacement of other regulatory factors (28). This may indeed be the case for the ovine PrP promoter, in which the YY1 site (motif 1) is adjacent to the E4BP4 site (motif 2). However our data from the tissue-derived nuclear extracts suggest that binding factors other than YY1 may act in a similar role. The two YY1-independent protein complexes (here designated Fy-I and Fy-II (NE proteins of unknown identity)) that were observed with the brain nuclear extracts will most likely compete for binding with YY1 in a similar way to the E4BP4 competition with other PAR factors. Our proposed regulatory model involving YY1 and E4BP4 (Fig. 6) may, therefore, be more intricate when other cells/tissue types are considered. Intriguingly, the discovery of complex II in brain extract from sheep and mouse indicates that wild type motif 1 can bind protein(s) specifically, and this may explain why motif 1 is conserved in all sequenced PrP promoters. Chromatin immunoprecipitation assays (29) could be used to provide a clearer picture of the role that YY1, E4BP4, and these other factors play in PrP gene expression in vivo.
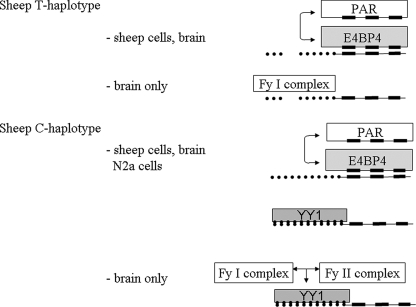
FIGURE 6.
Summary of the observed interactions of protein factors with PrP gene promoter C- and T-haplotypes. Our results suggest that the two ovine haplotypes are under different transcriptional control. The T-haplotype is activated by E4BP4 (or PAR) transcription factors in motif 2, whereas motif 1 forms protein complexes only with brain tissue extracts. The C-haplotype is equally activated by E4BP4 (or PAR) in motif 2 but is likely to be influenced in binding by YY1, which binds to motif 1. In brain tissue complex formation with other factors will compete with binding of YY1 in motif 1.
The E4BP4 Transcription Factor Binds to Motif 2—A species-specific C to T sequence variation of motif 2 has been described, where the ruminant sequence is CCATTACGTAACG and the non-ruminant sequence is CCATTATGTAACG (14). The ruminant motif 2 (M2C) probe formed a strong protein-DNA complex with a factor present in the ovine and murine N2a cell extracts (Fig. 3A), whereas the non-ruminant (M2T) probe formed a similar complex but with significantly lower affinity. The ruminant motif 2 sequence contains a consensus palindromic binding site (TTACGTAA) for the bZIP repressor protein E4BP4 (also known as nuclear factor, interleukin 3 (NFIL3)), which is a dimeric transcription factor comprised of two 52-kDa monomers (23). E4BP4 shares homology with the proline-acidic amino acid-rich (PAR) family of bZIP transcription factors (23). It has been proposed that competition between E4BP4 and PAR factors for the same binding site could act as a form of rheostat switch (30). Mobility shift assays of the M2C probe with an anti-E4BP4 antibody and with murine N2a and human HeLa cell nuclear extracts, respectively, resulted in supershift, indicating the binding of E4BP4 to this probe (Fig. 3C). Ovine nuclear extracts failed to produce a supershift or blocking reaction (Fig. 3B), which may be due to sequence changes in the epitope of ovine E4BP4; the antibody has not been tested with purified ovine E4BP4.6
Although it cannot be formally excluded that another factor bound to motif 2 in the ovine nuclear extracts, several reasons argue against this. First, the murine E4BP4-M2C DNA-protein complex showed very similar mobility to the complex found in ovine pA80BR nuclear extract. Second, the consensus binding site for E4BP4 is the palindromic sequence (TTACGTAA), and this is very common in dimeric transcription factor binding sites, as each monomer will bind to one identical half-site (bottom strand 3′-AATG-5′ and top strand 5′-GTAA-3′) (31). The differential affinity of the ovine nuclear protein for the TTA(C/T)GTAA sequence is, therefore, typical for homodimeric E4BP4. Third, the introduction of a methylated C residue into the M2C probe (TTA(mC)GTAA) did not alter the affinity of the protein in the ovine nuclear extracts (data not shown). C-5-methylated cytosine resembles thymidine in structure, and it would, therefore, be expected that the methylated probe may have a similar effect on protein binding as the non-ruminant M2T probe (TTATGTAA). As this was not the case, it is reasonable to assume that the binding is as expected for E4BP4 to the bottom strand 3′-AATG-5′, which is not modified by methylation but differs to the non-ruminant sequence 3′-AATA-5′. This also means that CpG methylation of motif 2 in the ovine promoter DNA is unlikely to affect gene regulation by E4BP4. The E4BP4 transcription factor has been shown to play a pivotal role in the regulation of circadian rhythms (23, 32). This is of interest in light of the observation that PrPc itself may exhibit a role in circadian rhythms as shown for prn-p knock-out mice (33) and suggested by the symptoms of fatal familial insomnia (FFI), a prion disease in humans in which patients initially suffer from interrupted sleep patterns (34). In fact, prn-p mRNA has been shown to be regulated in a circadian manner (35). The observation that E4BP4, which has defined roles in the regulation of circadian rhythms, interacts specifically with the PrP gene promoter may prove important for the understanding of the normal function of PrPc and the mechanisms of TSE pathogenesis and susceptibility. Moreover, E4BP4 has been shown to have defined roles in the control of apoptosis and in signal transduction pathways (36, 37), both of which are implicated in the pathogenesis of prion diseases (38, 39).
The reporter gene assay with construct p6TC in N2A cells suggests that E4BP4 acts as activator in the context of the PrP promoter, and this activity can be suppressed by YY1. In this model E4BP4 and YY1 appear to have an equal chance to bind to their respective motifs, and each cell either follows E4BP4 activation or YY1 repression, so that on average over all cells the effect is equal to the construct without E4BP4 and YY1 binding (construct p110-1 versus construct pTAA, Fig. 5C). Whether this is in fact the situation in tissues remains to be seen. Our data strongly support a role for E4BP4 in the regulation of the PrP gene; however, they do not exclude a role for other PAR factors in mediating these effects. We intend to follow this up in future studies.
Conclusion—Our data demonstrate that at the cellular level PrP transcription is regulated by transcription factors that have been shown to be involved in neurodegeneration (YY1) and periodic time regulation (E4BP4). How might this cellular mechanism translate into processes that influence the susceptibility to and pathogenesis of TSE disease in a sheep? Although it is difficult to predict how major a role this regulation plays, recent studies by Dupre et al. (40) emphasized that PrP was one of the strongest photoperiodically regulated genes in the pituitary gland. Temporal variation of PrP expression might be another as yet poorly understood mechanism involved in TSE infectivity spread. Recently it was shown in mouse and cellular models that there is a link between Alzheimer disease progression and PrP expression (41, 42). YY1 was reported to act as an activator of the BACE1 (β-site amyloid precursor protein-cleaving enzyme 1) gene promoter (43); it also increases transcription of protein FE65, which is involved in the processing and trafficking of amyloid precursor protein APP (44). In contrast, repression by YY1 of PrP expression could be seen as a counter-mechanism in amyloid precursor protein processing additional to the significant effects it could have on the development of TSE. Importantly, this regulation is haplotype- and species-specific, which means that there might be genetic subgroups with differing degrees of neurodegeneration in these diseases based on PrP promoter genotype. Further genetic studies in sheep scrapie but also in human TSEs such as fatal familial insomnia (45) may reveal the real importance of these PrP promoter motifs.
In summary, two motifs in the PrP promoter were previously defined purely on the basis of their conservation between species. Our data prove for the first time that protein binding can be ascribed to these motifs. At least four different protein complexes were observed, of which we identified two as being the transcription factors YY1 and E4BP4. The ovine motif 1 polymorphism and the PrP promoter sequence variation among species, i.e. between the TSE model organism mouse and the natural TSE host sheep, have provided an intricate regulatory model for PrP expression suitable for further molecular studies.
Acknowledgments
We thank Dr. Herbert Baybutt, The Roslin Institute, and Dr. Jim Allan, Institute of Cell Biology, University of Edinburgh, for experimental help and comments on the manuscript and Elliot Armstrong, Roslin Institute, for assistance with the figures.
This work was supported by United Kingdom Biotechnology and Biological Sciences Research Council (BBSRC) Grant 00A1D06533, Roslin ISPG Programme 3 “The Biology of Neurodegenerative Disease,” and BBSRC China Partnering Award CPA1741. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PrPc, cellular prion protein; TSE, transmissible spongiform encephalopathy; YY1, Yin Yang 1 transcription factor; bZIP, basic leucine zipper transcription factor; EMSA, electrophoretic mobility shift assay; NE, nuclear extract; PAR, proline-acidic amino acid-rich transcription factors.
S. T. G. Burgess, C. Shen, L. A. Ferguson, G. T. O'Neill, K. Docherty, N. Hunter, and W. Goldmann, unpublished results.
Santa Cruz Biotechnology, personal communication.
References
- 1.Weissmann, C., Aguzzi, A., Dormont, D., and Hunter, N. (2003) in British Medical Bulletin Series of Expert Reviews, pp. 321, Oxford University Press, Oxford
- 2.Prusiner, S. B. (1982) Science 216 136–144 [DOI] [PubMed] [Google Scholar]
- 3.Manson, J. C., Clarke, A. R., Hooper, M. L., Aitchison, L., McConnell, I., and Hope, J. (1994) Mol. Neurobiol. 8 121–127 [DOI] [PubMed] [Google Scholar]
- 4.Manson, J. C., Clarke, A. R., McBride, P. A., McConnell, I., and Hope, J. (1994b) Neurodegeneration 3 331–340 [PubMed] [Google Scholar]
- 5.Johnson, R. T. (2005) Lancet Neurol. 10 635–642 [DOI] [PubMed] [Google Scholar]
- 6.Hu, W., Rosenberg, R. N., and Stüve, O. (2007) Acta Neurol. Scand. 116 75–82 [DOI] [PubMed] [Google Scholar]
- 7.Zomosa-Signoret, V., Arnaud, J. D., Fontes, P., Alvarez-Martinez, M. T., and Liautard, J. P. (2008) Vet. Res. 39 9. [DOI] [PubMed] [Google Scholar]
- 8.Schneider, K., Korkmaz, Y., Addicks, K., Lang, H., and Raab, W. H. (2007) J. Endod. 33 110–113 [DOI] [PubMed] [Google Scholar]
- 9.Saeki, K., Matsumoto, Y., Matsumoto, Y., and Onodera, T. (1996) Biochem. Biophys. Res. Commun. 219 47–52 [DOI] [PubMed] [Google Scholar]
- 10.Inoue, S., Tanaka, M., Horiuchi, M., Ishiguro, N., and Shinagawa, M. (1997) J. Vet. Med. Sci. 59 175–183 [DOI] [PubMed] [Google Scholar]
- 11.Haigh, C. L., Wright, J. A., and Brown, D. R. (2007) J. Mol. Biol. 368 915–927 [DOI] [PubMed] [Google Scholar]
- 12.Premzl, M., and Gamulin, V. (2007) BMC Genomics 8 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sander, P., Hamann, H., Drögemüller, C., Kashkevich, K., Schiebel, K., and Leeb, T. (2005) J. Biol. Chem. 280 37408–37414 [DOI] [PubMed] [Google Scholar]
- 14.Westaway, D., Cooper, C., Turner, S., Dacosta, M., Carlson, G. A., and Prusiner, S. B. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 6418–6422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Neill, G. T., Donnelly, K., Marshall, E., Cairns, D., Goldmann, W., and Hunter, N. (2003) J. Anim. Breed Genet. 120 114–123 [Google Scholar]
- 16.O'Neill Cairns, D., Goldmann, W., Toovey, L., and Hunter, N. (2005) J. Anim. Breed. Genet. 122 86–94 [DOI] [PubMed] [Google Scholar]
- 17.John, H. A. (1994) Hum. Gene Ther. 5 283–293 [DOI] [PubMed] [Google Scholar]
- 18.Freshney, R. I. (1987) Culture of Animal Cells; A Manual of Basic Technique, 2nd Ed., Wiley-Liss, Inc., New York
- 19.Wingender, E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Pruss, M., Reuter, I., and Schacherer, F. (2000) Nucleic Acids Res. 28 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi, Y., Lee, J. S., and Galvin, K. M. (1997) Biochim. Biophys. Acta 1332 49–66 [DOI] [PubMed] [Google Scholar]
- 21.Johansson, E., Hjortsberg, K., and Thelander, L. (1998) J. Biol. Chem. 273 29816–29821 [DOI] [PubMed] [Google Scholar]
- 22.Dyer, K. D., and Rosenberg, H. F. (2001) Life Sci. 69 201–212 [DOI] [PubMed] [Google Scholar]
- 23.Cowell, I. G. (2002) BioEssays 24 1023–1029 [DOI] [PubMed] [Google Scholar]
- 24.Hyde-DeRuyscher, R. P., Jennings, E., and Shenk, T. (1995) Nucleic Acids Res. 23 4457–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yant, S. R., Zhu, W., Millinoff, D., Slightom, J. L., Goodman, M., and Gumucio, D. L. (1995) Nucleic Acids Res. 23 4353–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romey, M. C., Pallares-Ruiz, N., Mange, A., Mettling, C., Peytavi, R., Demaille, J., and Claustres, M. (2000) J. Biol. Chem. 275 3561–3567 [DOI] [PubMed] [Google Scholar]
- 27.Seto, E., Lewis, B., and Shenk, T. (1993) Nature 365 462–464 [DOI] [PubMed] [Google Scholar]
- 28.Ericsson, J., Usheva, A., and Edwards, P. A. (1999) J. Biol. Chem. 274 14508–14513 [DOI] [PubMed] [Google Scholar]
- 29.Spencer, V. A., Sun, J. M., Li, L., and Davie, J. R. (2003) Methods 31 67–75 [DOI] [PubMed] [Google Scholar]
- 30.Ikushima, S., Inukai, T., Inaba, T., Nimer, S. D., Cleveland, J. L., and Look, A. T. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 2609–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKnight, S. L. (1991) Sci. Am. 264 54–64 [DOI] [PubMed] [Google Scholar]
- 32.Mitsui, S., Yamaguchi, S., Matsuo, T., Ishida, Y., and Okamura, H. (2001) Genes Dev. 15 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tobler, I., Gaus, S. E., Deboer, T., Achermann, P., Fischer, M., Rulicke, T., Moser, M., Oesch, B., McBride, P. A., and Manson, J. C. (1996) Nature 380 639–642 [DOI] [PubMed] [Google Scholar]
- 34.Gambetti, P., Petersen, R., Monari, L., Tabaton, M., Autiliogambetti, L., Cortelli, P., Montagna, P., and Lugaresi, E. (1993) Br. Med. Bull. 49 980–994 [DOI] [PubMed] [Google Scholar]
- 35.Cagampang, F. R. A., Whatley, S. A., Mitchell, A. L., Powell, J. F., Campbell, I. C., and Coen, C. W. (1999) Neurosci. 91 1201–1204 [DOI] [PubMed] [Google Scholar]
- 36.Nishimura, Y., and Tanaka, T. (2001) J. Biol. Chem. 276 19921–19928 [DOI] [PubMed] [Google Scholar]
- 37.Yu, Y. L., Chiang, Y. J., and Yen, J. J. (2002) J. Biol. Chem. 277 27144–27153 [DOI] [PubMed] [Google Scholar]
- 38.Jamieson, E., Jeffrey, M., Ironside, J. W., and Fraser, J. R. (2001) Neuroreport 12 3567–3572 [DOI] [PubMed] [Google Scholar]
- 39.Kretzschmar, H. A., Giese, A., Brown, D. R., Herms, J., Keller, B., Schmidt, B., and Groschup, M. (1997) J. Neural Transm. Suppl. 50 191–210 [DOI] [PubMed] [Google Scholar]
- 40.Dupre, S. M., Burt, D. W., Talbot, R., Downing A., Mouzaki, D., Waddington, D., Malpaux, B., Davis, J. R. E., Lincoln, G. A., and Loudon, A. S. I. (2008) Endocrinology 149 5527–5539 [DOI] [PubMed] [Google Scholar]
- 41.Parkin, E. T., Watt, N. T., Hussain, I., Eckman, E. A., Eckman, C. B., Manson, J. C., Baybutt, H. N., Turner, A. J., and Hooper, N. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11062–11067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baier, M., Apelt, J., Riemer, C., Gültner, S., Schwarz, A., Bamme, T., Burwinkel, M., and Schliebs, R. (2008) Int. J. Dev. Neurosci. 26 821–824 [DOI] [PubMed] [Google Scholar]
- 43.Novak, K., Lange-Dohna, C., Zeitschel, U., Gunther, A., Luscher, B., Robitzki, A., Preez-Polo, R., and Rossner, S. (2006) J. Neurochem. 96 1696–1707 [DOI] [PubMed] [Google Scholar]
- 44.Zambrano, N., De Renzis, S., Minopoli, G., Faraonio, R., Donini, V., Scaloni, A., Cimino, F., and Russo, T. (1997) Biochem. J. 328 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montagna, P. (2005) Sleep Med. Rev. 5 339–353 [DOI] [PubMed] [Google Scholar]