TABLE 2.
Refinement statistics and contents in the asymmetric unit
| Crystal | Ligand-free | GalNAc | GlcNAc | GlcNAc-NO3-EG | GlcNAc-SO4 |
|---|---|---|---|---|---|
| Refinement statistics | |||||
| Protein Data Bank code | 2ZUS | 2ZUT | 2ZUU | 2ZUV | 2ZUW |
| Resolution range (Å) | 32.1-2.11 | 44.8-1.90 | 40.7-2.30 | 35.1-1.85 | 41.1-2.11 |
| R-factor/Rfreea (%) | 17.0/22.5 | 16.1/20.4 | 17.0/23.1 | 14.9/19.2 | 16.9/22.5 |
| r.m.s. deviation bond lengths (Å) | 0.014 | 0.014 | 0.010 | 0.014 | 0.013 |
| r.m.s. deviation bond angles (degrees) | 1.3 | 1.4 | 1.2 | 1.3 | 1.3 |
| Average B-factor (Å2) | |||||
| Protein (chain A/B/C/D) | 25.4/30.9/26.8/27.5 | 24.7/30.4/24.8/28.4 | 32.1/39.0/31.9/37.2 | 20.6/20.2 | 22.3/28.7/22.9/27.6 |
| Ligands in each active site | - | 22.2/24.0/21.3/23.0 | 26.2/32.7/29.3/30.5 | 20.0/14.6 | 19.2/22.6/20.0/34.2 |
| Water | 31.5 | 36.0 | 37.1 | 33.6 | 32.4 |
| Ramachandran plot (% in favored/allowed/disallowed regions) | |||||
| Chain A | 97.1/2.3/0.7 | 96.6/3.0/0.4 | 96.5/3.1/0.4 | 97.3/2.6/0.1 | 96.8/3.2/0 |
| Chain B | 96.2/3.4/0.4 | 96.4/3.6/0 | 94.9/4.6/0.6 | 96.6/3.3/0.1 | 95.9/3.7/0.4 |
| Chain C | 96.5/3.0/0.5 | 97.0/2.8/0.1 | 96.6/3.4/0 | 96.5/3.4/0.1 | |
| Chain D | 96.6/3.4/0 | 96.5/3.4/0.1 | 95.4/4.5/0.1 | 95.1/4.3/0.6 | |
| ASU content | |||||
| No. of protein atoms | 23,724 | 23,573 | 23,549 | 11,787 | 23,395 |
| No. of solvent atoms | 1,898 | 3,114 | 1,638 | 1,826 | 2,281 |
| No. of heteroatoms | 2 | 124 | 85 | 94 | 106 |
| No. of ligands | |||||
| Mg2+ | 2 | 6 | 1 | 8 | 1 |
| GalNAc/GlcNAc | 4 (GalNAc) | 4 (GlcNAc) | 2 (GlcNAc) | 4 (GlcNAc) | |
| Glycerol/EG | 9 (glycerol) | 4 (glycerol) | 9 (EG) | 6 (glycerol) | |
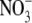 / /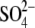
|
1 (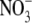 ) )
|
1 (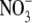 ) )
|
1 (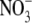 ) + 1
( ) + 1
(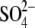 ) )
|
||
| Model built/Conformationb/Ligands in active site | |||||
| Chain A | 2-750/open/NAc | 3-34, 43-754/semiclosed/GalNAc | 3-34, 41-754/semiclosed/GlcNAc | 3-35, 42-749/closed/GlcNAc, EG, 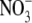
|
3-33, 40-753/semiclosed/GlcNAc |
| Chain B | 3-34, 37-41, 43,44, 47-751/open/NA | 3-34, 47-749/semiclosed/GalNAc | 3-33, 47-367, 371-749/semiclosed/GlcNAc | 3-33, 42-750/semiclosed/GlcNAc | 3-32, 40-366, 371-751/semiclosed/GlcNAc |
| Chain C | 3-35, 38-40, 43-750/open/NA | 3-33, 43-754/semiclosed/GalNAc | 3-34, 43-754/semiclosed/GlcNAc | 3-35, 40-754/semiclosed/GlcNAc | |
| Chain D | 3-35, 39-750/open/NA | 334, 43750/semiclosed/GalNAc | 3-33, 43-750/semiclosed/GlcNAc |
5-12, 22-33, 46-372, 375-400, 407-427, 433-751/closed/GlcNAc,
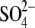
|
Rfree = ∑hkl||Fo| - |Fc||/∑hkl|Fo|, where the crystallographic R-factor was calculated including and excluding refinement reflections. The free reflections constituted 5% of the total number of reflections.
See “Results” for designation of conformations.
NA, not applicable.
