Abstract
A prominent feature of most if not all cancers is a striking genetic instability, leading to ongoing accrual of mutational changes, some of which underlie tumor progression, including acquisition of invasiveness, drug resistance, and metastasis. Thus, the molecular basis for the generation of this genetic diversity in cancer cells has important implications in understanding cancer progression. Here we report that homologous recombination (HR) activity is elevated in multiple myeloma (MM) cells and leads to an increased rate of mutation and progressive accumulation of genetic variation over time. We demonstrate that the inhibition of HR activity in MM cells by small inhibitory RNA (siRNAs) targeting recombinase leads to significant reduction in the acquisition of new genetic changes in the genome and, conversely, the induction of HR activity leads to significant elevation in the number of new mutations over time and development of drug resistance in MM cells. These data identify dysregulated HR activity as a key mediator of DNA instability and progression of MM, with potential as a therapeutic target.
Introduction
Genetic changes observed at the chromosomal level or at the nucleotide sequence level are associated with the development and progression of malignant phenotypes. Although some specific cancers are associated with and attributed to specific cytogenetic and molecular aberrations, for example, chronic myelogenous leukemia or acute promyelocytic leukemia, the majority of cancers display a complex spectra of diverse genetic alterations apparent at diagnosis and acquire additional changes with progression of the disease.1 Because the large-scale chromosomal alterations that arise frequently in cancer cells occur infrequently in normal cells,2–4 it implies that the control mechanisms that maintain the integrity of chromosomes in normal cells are disrupted in cancer cells.
A variety of intrinsic or extrinsic chemical as well as physical factors damage DNA in living organisms which, if not repaired, can lead to mutations and cellular transformation. The best known machinery involved in repairing potentially lethal DNA damage, especially double-strand breaks, is genetic recombination.5 In fact the repair of DNA lesions may account for majority of the recombination occurring in mitosis.6
Recombination plays an important role in maintaining the genetic integrity of a cell, including DNA repair7 and proper segregation of chromosomes in meiosis.8 In the normal cellular environment, recombination-associated proteins are highly regulated, precise, and exhibit considerable specificity for DNA sequences, which have either an extensive homology or a characteristic signal sequence. However, with its significant ability to modify DNA, if dysregulated, it can lead to genomic instability. Recombination can be induced by several chemicals, radiation, and oncogenic viruses.9–14 The induction or overexpression of a recombination pathway may result in DNA rearrangements, leading to oncogene activation15 and/or the loss of hemizygous functional alleles of tumor suppressor genes.
Aberrant or dysregulated recombination has been implicated in chromosome translocation,9,16,17 gene amplification,18 and telomere maintenance19 and may therefore underlie the chromosomal aberrations observed with high frequency in many cancers. Previous studies, from our group and others, have implied an important role of homologous recombination (HR) in the process leading to cellular transformation.1,9,16,17,20–25
In this study, we used multiple myeloma (MM) as a model cancer to evaluate the molecular mechanisms of acquisition of genetic instability and progression of cancer because significant chromosomal instability is observed in this malignancy, with more than 11 cytogenetic abnormalities observed per karyotype and with demonstrated progressive cytogenetic change over time in patient samples and in myeloma-derived cell lines.26–28 Here we report that HR activity is constitutively elevated in MM cell lines and patient samples and demonstrate that the inhibition of HR activity leads to a significant reduction in the acquisition of new genetic changes whereas induction of HR activity leads to significant elevation in the number of new mutations over time and development of drug resistance in MM cells.
Methods
Myeloma and normal cells
MM cell lines RPMI8226 and U266 were obtained from the ATCC (Rockville, MD); MM lines ARD, ARK, and ARP1 were kindly provided by Dr J. Epstein (University of Arkansas for Medical Sciences, Little Rock, AR), and MM1S was kindly provided by Dr Steven Rosen (Northwestern University, Chicago, IL). All cell lines were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (HyClone, South Logan, UT), as described previously29–32 and were maintained in a state of logarithmic growth. For RNA extraction, cultures were harvested at the same final cell density (5 × 105/mL) and immediately processed.
In accordance with the Declaration of Helsinki and institutional research board approval from Dana-Farber Cancer Institute, myeloma cells were isolated from bone marrow aspirate samples that were obtained with informed consent from patients with MM by positive immunomagnetic bead selection with the use of anti-CD138 antibodies and magnet-assisted cell sorting (MACS; Miltenyi Biotec, Auburn, CA), according to the manufacturer's instructions. We assessed the purity of plasma cells (> 95%) by monitoring cell-surface expression of CD38 and CD45.33
Recombination assay
HR frequency was measured with the use of a plasmid substrate as described previously.21,25 Normal and MM cells were transfected with a plasmid substrate for recombination, DR1.34 After 36 hours, plasmid DNA was recovered, introduced into competent (RecA−) Escherichia coli (Invitrogen, Carlsbad, CA), and plated on Luria-Bertani medium (LB)–agar plates with either ampicillin (to count total plasmid-transfected colonies) or ampicillin plus neomycin (to count recombined plasmids in which an intact neomycin resistance gene had been generated by recombination). After 16 hours, bacterial colonies were counted and the recombination frequency was calculated from the ratio of these 2 counts.
Gene expression analysis and biostatistics
Total RNA was isolated with an RNeasy kit (QIAGEN, Valencia, CA) and gene expression profile was evaluated with HG-U133 array (Affymetrix, Santa Clara, CA) representing approximately 33 000 human genes as described previously.30,35–38 GeneChip arrays were scanned on a GeneArray Scanner (Affymetrix). Array normalization, expression value calculation, and clustering analysis were performed by the use of the dChip Analyzer. The invariant set normalization method was used to normalize arrays at probe level to make them comparable, and the model-based method was used for probe selection and to compute expression values.39,40 These expression levels were assigned standard errors based on replicates, which were subsequently used to compute 90% confidence intervals of fold changes in intergroup comparisons. The lower confidence bounds of fold change are conservative estimates of the actual changes.
Immunocytochemical detection of HsRAD51 and paralogs
Cytospins of normal plasma cells and MM cells were fixed in methanol/acetone (1:1, vol/vol) for 10 minutes at −20°C. Fixed cells were rinsed, rehydrated in phosphate-buffered saline (PBS), and incubated for 2 hours at room temperature (RT) with rabbit polyclonal antibody to HsRAD51 or mouse monoclonal antibodies to HsRAD51B, HsRAD51C, HsRAD51D, XRCC2, and XRCC3 (Novus Biologicals, Littleton, CO). Immunostaining of RAD51 and paralogs were performed with the Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA), as described by the manufacturer.
Small inhibitory RNAs and transfections
We designed 2 small inhibitory RNA (siRNA) duplexes targeting different regions of the HsRAD51 gene according to Harborth's criteria41 for choosing sequences and making siRNA duplexes for target mRNAs. SiRNAs with sense-strand sequences 5′-AAAGAGCTTGACAA-ACTACTT-3′ and 5′-AACTGGGAAGACCCAGATCTG-3′ were submitted in a BLAST search of human EST libraries to ensure that other human genes were not targeted. Preannealed siRNA duplexes of the aforementioned sequences were purchased from Dharmacon Research (Lafayette, CO). SiRNA duplexes were transfected into MM cells by use of the TransIT-TKO Transfection Reagent (Mirus, Madison, WI), as described by the manufacturer. In brief, cells were plated in complete growth medium 24 hours before transfection, at 2 × 105/mL, and incubated overnight. Immediately before transfection, TransIT-TKO Reagent was added dropwise to serum-free medium (RPMI 1640) and incubated at RT for 20 minutes. SiRNA duplexes (25 nmol/L each) were added to diluted TransIT-TKO reagent, mixed, and incubated at RT for 20 minutes. SiRNA-TKO complexes were then layered dropwise onto the cells and incubated as described.41,42
Promoter regulation of HsRAD51
HsRAD51 promoter was cloned upstream of a luciferase gene in a mammalian expression vector. The construct (HsRAD51P-LUC) was transfected into normal diploid fibroblasts, and the cells were then exposed to nickel chloride. After a 2-hour exposure, cells were lysed and luciferase activity was assayed with a Luciferase Assay Kit (Clontech, Mountain View, CA).
Genomewide loss of heterozygosity
We assayed genomewide loss of heterozygosity (LOH) as described (Affymetrix) to estimate genomic instability and ongoing DNA rearrangements. The HuSNP and 10K arrays (Affymetrix) allow human genomewide monitoring of approximately 1500 and 10 000 loci, respectively. Genomic DNA was isolated from myeloma cells cultured in the presence of recombination inhibitors or inducer, or without any addition, and polymorphic loci were amplified by multiplex polymerase chain reaction (PCR). PCR products were pooled, labeled, and hybridized to HuSNP or 10K probe arrays according to the manufacturer's directions. Normalization of arrays and analyses of data were performed with the DNA-Chip Analyzer (dChip) program.40,43 Genotype calls from cell samples harvested and frozen at the beginning of experiment (day 0) were used as baselines to identify new LOH loci in the treated cells.
Results
Homologous recombination activity is significantly elevated in MM cells
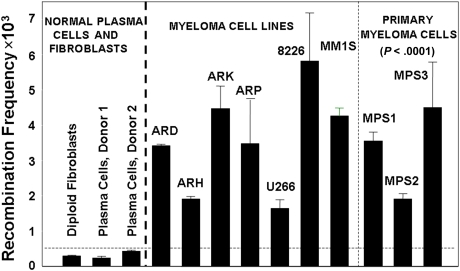
To investigate the levels of HR activity in MM cells, we used a plasmid-based assay for homologous recombination.21,25 We transfected diploid human fibroblasts, normal plasma cells from healthy donors, 6 MM cell lines, and malignant plasma cells purified from bone marrow of 3 different MM patients, with a recombination substrate plasmid.21,25 After 36 hours, plasmid DNA was recovered from these cells and the generation of a functional neomycin resistance gene, as a measure of recombination activity, was determined. As shown in Figure 1, a significant elevation in recombination activity was observed in myeloma cell lines and primary myeloma cells, averaging 11-fold greater than the mean value obtained for normal cells (range, 5- to >18-fold; P < .0001). Significant elevation of HR activity also was observed in Barrett associated adenocarcinoma, another malignancy with pronounced and varied cytogenetic abnormality (data not shown).
Figure 1.
Increased HR activity in MM cells. HR activity was measured with the recombination plasmid substrate. Generation of the neomycin resistance as a measure of recombination activity was evaluated by counting neomycin-resistant colonies and total plasmid-transfected was evaluated by ampicillin-resistant colonies. The recombination frequency was calculated from the ratio of these 2 counts. Recombination frequency is presented as number of recombination events per million transfected plasmid copies.
Transcript and protein levels of recombinase and related genes are elevated in myeloma
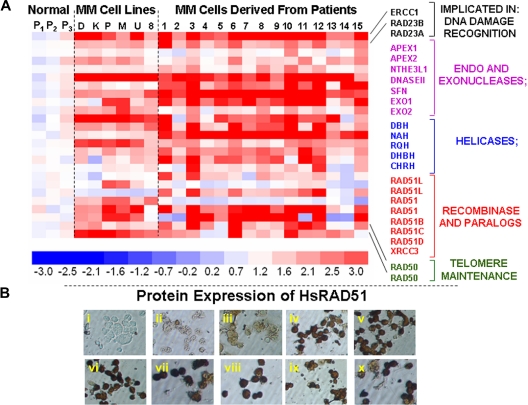
To identify genes that may be implicated in the elevated HR activity and genomic instability in multiple myeloma, we evaluated a genome-wide expression profile of 6 myeloma cell lines, 15 myeloma patient samples, and 3 normal plasma cell isolates. We noted significant up-regulation of a subset of known recombination-related genes in MM cell lines and patient samples, relative to normal plasma cells (Figure 2A). These expression results were reproducible between experiments (r = 0.99). Transcript levels of number of HR-related genes, including endo- and exonucleases, helicases, HsRAD51D (implicated in homologous pairing, strand exchange and telomere maintenance), HsRAD51 paralog XRCC3, and RAD50 (implicated in double-strand break recognition/signaling and telomeric recombination), were significantly elevated in MM cell lines and primary cells (Figure 2A). Immunocytochemistry confirmed a marked up-regulation of these proteins in various MM cell lines, of which a representative example is shown in Figure 2B.
Figure 2.
Elevated expression of HR-related genes in MM cells. (A) Gene expression profile was measured in 3 normal plasma cell isolates (P1, P2, P3), 6 myeloma cell lines (D indicates ARD; K, ARK; P, ARP; M, MM1s; U, U266; and 8, RPMI8226), and 15 purified myeloma cells from patient bone marrows (1-15) with the use of HG-U133 gene arrays. The color scale at the bottom of the figure represents fold change in recombination- and repair-associated genes in myeloma cells relative to average expression in 3 normal plasma cell samples. (B) Protein levels of HsRAD51 and its paralogs were detected by immunostaining, with either a polyclonal antibody against HsRAD51, also recognizing RAD51B and RAD51C or monoclonal antibodies recognizing HsRAD51 or its paralogs individually. (i) Normal plasma cells, no primary antibody control; (ii) normal plasma cells (donor 1), HsRAD51/51B/51C; (iii) normal plasma cells (donor 2), HsRAD51/51B/51C; (iv) ARP myeloma cell line, HsRAD51/51B/51C; (v) MM1S myeloma cell line, HsRAD51/51B/51C; (vi) U266 myeloma cell line, HsRAD51/51B/51C; (vii) patient myeloma cells, HsRAD51; (viii) patient myeloma cells HsRAD51B; (ix) patient myeloma cells, HsRAD51C; and (x) patient myeloma cells, HsRAD51D.
Myeloma cells acquire new genetic alterations over time
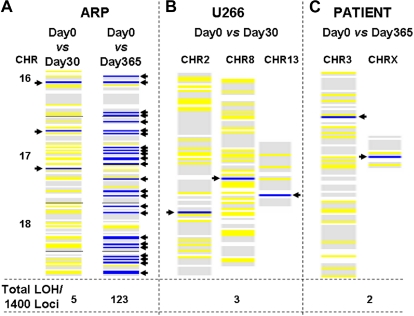
We investigated whether MM cells with elevated HR activity continue to acquire new genetic alterations over time. We used a genomewide LOH assay44 based on single nucleotide polymorphism (SNP) genotyping with Affymetrix chips as a tool to estimate the rate of mutation and genomic instability. MM cells in culture were harvested at several time points, and the genomic DNA was analyzed with HuSNP arrays, which allow interrogation of approximately 1400 genome-wide loci. Using the genotype calls from cell samples harvested at the beginning of an experiment (day 0) as a baseline, ARP cells acquired 5 and 123 new regions with LOH during 1 and 12 months in culture, respectively (Figure 3A); similarly, we observed 3 new LOH regions during a period of 30 days in U266 cells (Figure 3B). Bone marrow samples collected from a myeloma patient before and after a 12-month interval also revealed 2 new LOH loci per 1400 sites examined in the genome (Figure 3C). Similarly, the myeloma cells were collected from 4 additional patients at 6-month intervals and evaluated for acquisition of new genomic changes with the use of 500k SNP arrays. In all 4 patients, we observed the acquisition of new genomic changes identified as new LOH increasing in number with time (data not shown), providing evidence of recombination or gene rearrangement events occurring in vivo.
Figure 3.
Acquisition of new genomic changes in multiple myeloma cells over time. Myeloma cells were cultured for various durations. Aliquots of cells were removed at specified intervals and genomic DNA was isolated and evaluated for LOH with the use of HuSNP chips (Affymetrix). DNA from the cells harvested and frozen at day 0 was used to define the allelotype baseline, departures from which identified new LOH loci in cells harvested at later time points. Blue lines show new LOH loci in the regions of genome indicated, whereas yellow lines represent retention of preexisting LOH sites. The numbers at the bottom of each figure indicate total number of new LOH loci throughout genome, per 1400 sites investigated. (A) New LOH loci in ARP myeloma cells cultured for either 1 or 12 months, in the region of genome spanning chromosomes 16, 17, and 18, are shown as an example. (B) New LOH loci in U266 myeloma cells cultured for 1 month. (C) New LOH loci in genomic DNA samples, derived from a patient at 1-year intervals.
Effect of inhibition and augmentation of HR activity on myeloma cells
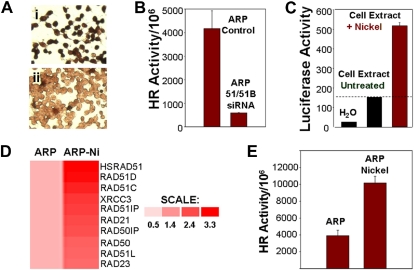
We investigated whether induction or suppression of components of the HR pathway exert effects on HR activity and genomic instability. A siRNA, directed against both HsRAD51 and HsRAD51B, was transfected into ARP myeloma cells with elevated transcript levels of HsRAD51 (Figure 2A). Transfection with HsRAD51 siRNA (transfection efficiency > 95%) was associated with loss of HsRAD51 protein expression in 86% plus or minus 4% of the cells (Figure 4Aii), relative to cells transfected with control siRNA (Figure 4Ai), and 88% plus or minus 3% inhibition of HR activity (Figure 4B). Exposure to nickel chloride, which we had previously shown to be a potent recombinogen,11 led to a 3.5-fold increase in the promoter activity of HsRAD51 recombinase (Figure 4C); 2- to 3-fold induction in the transcript levels of recombinase and related genes, including Rad51C, Rad51D, XRCC3, Rad23, and Rad50 (Figure 4D); and a further 2-fold stimulation of HR activity in myeloma cells (Figure 4E).
Figure 4.
HR activity in myeloma cells is induced by nickel chloride and inhibited by siRNAs targeting recombinases. (A) Loss of RAD51 protein expression by siRNAs. Immunostaining using monoclonal antibody to HsRAD51 of ARP cells transfected with (I) control siRNA and (II) HsRAD51-specific siRNAs. (B) Loss of HR activity by siRNAs. HR activity was assessed using the plasmid substrate after the transfection of ARP cells with siRNA directed at HsRAD51 and RAD51B. Recombination frequency is presented as number of recombination events per million transfected plasmid copies. ARP control, MM cells transfected with control siRNAs; ARP 51/51B SiRNA, MM cells transfected with siRNAs directed at RAD51 ands RAD51B. (C) Effect of nickel on promoter regulation of HsRAD51. HsRAD51 promoter was cloned upstream of a luciferase gene in a mammalian expression vector. The resulting construct HsRAD51P-LUC was transfected into normal diploid fibroblasts and the cells were then exposed to nickel chloride (0.3 mg/mL). After a 2-hour exposure, cells were lysed and luciferase activity was assayed with a Luciferase Assay Kit (Clontech). Lanes show luciferase activity in water (lane 1), in ARP cells transfected with RAD51P-Luc (lane 2), and in ARP cells transfected with RAD51P-Luc and exposed to nickel chloride (lane 3). (D) Induction of HsRAD51 and related genes by nickel chloride in ARP cells. ARP myeloma cells, untreated (ARP) or treated with nickel chloride (0.3 mg/mL) for 24 hours (ARP-Ni) were harvested, total RNA was isolated, and processed for gene expression analysis using U133 arrays (Affymetrix). Gene expression is shown by intensity of red color. Color scale shows fold change of gene expression in treated cells relative to untreated ARP cells. (E) HR activity in untreated ARP cells (ARP) or ARP cells exposed to nickel chloride (ARP Nickel; 0.3 mg/mL) for 5 days.
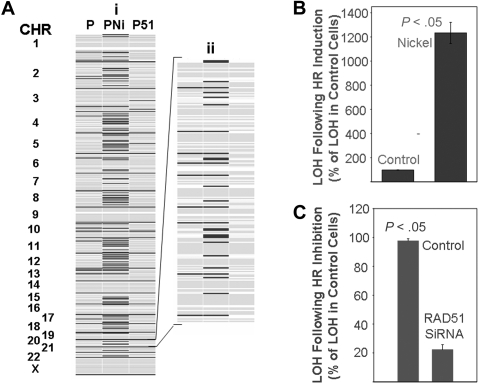
Next, we evaluated whether inhibition or induction of HR could affect the frequency of acquisition of new genetic changes in myeloma cells. We cultured myeloma cells (ARP) without any addition or in the presence of HsRAD51-siRNA or nickel chloride, as described in the figure legends. We evaluated genome-wide LOH by comparing genotypes before and after the treatment. In 3 independent experiments, inhibition of HR by HsRAD51-siRNA reduced the acquisition of new LOH loci by 78% plus or minus 3.6% (mean ± SEM; Figure 5A,C), whereas treatment of cells with nickel chloride increased the number of new LOH sites by more than 12-fold (Figure 5A,B).
Figure 5.
Effect of homologous recombination on rate of mutation in myeloma cells. (A) ARP myeloma cells were either transfected every 2 weeks with control siRNAs for 90 days (lane 1), exposed to nickel chloride (0.3 mg/mL) for 5 days (lane 2), or transfected every 2 weeks with siRNAs targeting HsRAD51 for 90 days (lane 3). Genomic DNA was isolated and evaluated with huSNP arrays (Affymetrix) as described. A representative figure showing all chromosomes (i) and an enlarged version of chromosome 20 (ii) is shown as an example. Dark lines indicate new LOH loci. (B,C) Summary of change in LOH over all chromosomes; error bars indicate standard error of the mean of triplicate assays. (B) Induction of LOH in ARP cells after exposure to nickel chloride as a percentage of control cell values. (C) Reduction in LOH in ARP cells after suppression of HR by HsRAD51 directed siRNA as a percentage of control cell values.
Homologous recombination mediates drug resistance in myeloma
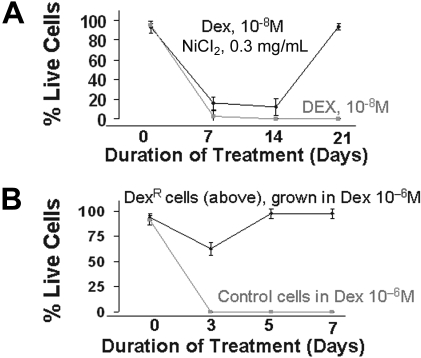
We next evaluated the effect of HR induction, and the consequent increase in genetic aberrations, on the development of drug resistance in MM. Myeloma cells were cultured in the presence of a cytotoxic dose of dexamethasone (10−8 mol/L), a potent drug used in the therapy of MM, with or without nickel chloride, a potent inducer of HR.11 Cell viability was measured weekly. As seen in Figure 6, no live cells were detected in cultures exposed to dexamethasone alone for 2 to 3 weeks, whereas in presence of nickel chloride, dexamethasone-resistant cells appeared to arise during the first week of exposure and predominated in the culture by day 21 (>95% live cells). Nickel chloride alone, at the same concentration, had no effect on cell viability (data not shown). To confirm the development of drug resistance, these resistant cells were exposed to a greater concentration of dexamethasone (10−6 mol/L) for 1 week. Although all the control cells were dead at day 3, no significant effects were observed on the resistant cells (Figure 6). These results recapitulate the observed in vivo development of drug resistance in patients after therapy and strongly implicate HR in this phenomenon.
Figure 6.
Induction of drug resistance by elevated HR activity. (A) ARP myeloma cells were cultured in the presence of dexamethasone (10−8 mol/L) alone or dex (10−8 mol/L) and nickel chloride (0.3 mg/mL). Live cell number was determined weekly by trypan blue staining. After 2 weeks of culture, nickel was removed from the dexamethasone and nickel-treated cultures. The figure is representative of 3 independent experiments. (B) Dexamethasone-resistant MM cells obtained in panel A were cultured in higher concentrations of dex (10−6 mol/L). Live cell number of control and dexamethasone-resistant ARP cells was determined by trypan blue staining.
Discussion
Genetic recombination, which occurs both in somatic tissues and in gametic lineages, plays a crucial role in repairing DNA damaged by a variety of origins. The process of recombination is well regulated and depends on a coordinated action of several proteins, including DNA damage recognizing/signaling proteins, topoisomerases, helicases, single-stranded DNA binding proteins, endonucleases, homologous base pairing (RAD51 and related) proteins, and DNA ligases. HsRAD51 recombinases are considered to be key players in HR. Rad51 proteins bind to both single- and double-stranded DNA and have both homologous-pairing and strand-exchange activities.45 To date, 5 HsRad51 paralogs have been identified: XRCC2, XRCC3 Rad51B/Rad51L1, Rad51C/Rad51L2, and Rad51D/Rad51L3,46 each with unique but overlapping mechanism of action providing redundancy to the system.
Although recombination-associated proteins are highly precise and exhibit considerable specificity for DNA sequences that have either an extensive homology or a characteristic signal sequence, their expression may be elevated or induced by extrinsic (carcinogens, radiation and oncogenic viruses)9–14 or intrinsic (oxidative metabolites of food) DNA damaging agents, leading to increased recombination activity. If homologous recombination becomes constitutively activated, it may cause genomic rearrangements and lead to oncogene activation15 and/or the loss of hemizygous functional alleles of tumor suppressor genes47 and may therefore facilitate the acquisition and progression of a tumor and/or drug-resistant phenotype.
Several HR-related genes are consistently up-regulated in MM (Figure 2) as well as other cancers,48–51 and these are presumed to exert a concerted action to produce the high HR activity observed in primary cancer cells and all cancer cell lines tested (Figure 2). These appear to comprise either a single, nonredundant pathway or else multiple pathways with common regulatory elements, in that inhibition of specific components (RAD51 or endonucleases) is sufficient to dramatically suppress HR and genetic instability (Figure 4). Additionally, up-regulation of many critical components of this pathway, observed in MM and other cancers, may underlie the striking increase in genetic instability characteristic of most, if not all, cancer cells. Consistent with this the elevated expression of RAD51, recombinase has been associated with a significantly shorter survival of patients with lung cancer.52 Similarly, BRCA1 and BRCA2, which are known as breast and ovarian cancer susceptibility proteins, have been implicated in the regulation of recombinational repair through their interaction with HsRAD51 recombinase.53 Mutations in these genes or in RAD51 itself are associated with aberrant recombinase activity, genomic instability, and development of breast cancer.
Although overexpression or mutations of HsRAD51 have been found in many cancers,48–51 the HR activity in these malignancies remains unexplored. Here we report increased HR activity and overexpression of several RAD51 paralogs and other HR-related transcripts and proteins in established MM cell lines as well as primary myeloma cells. We have confirmed our observation of elevated HR and expression of related genes in myeloma, in another malignancy, Barrett esophageal adenocarcinoma (data not shown), suggesting applicability of these findings in other cancers.
Because HR has been implicated in gene amplification18 and rearrangement,9,16,17,25 we evaluated whether cancer cells with elevated HR activity continue to acquire new genetic alterations over time. Genomewide analyses that use SNP arrays (Affymetrix),44 which allow investigation of more than 1400 LOH loci in the genome, could detect the acquisition of additional new mutations in myeloma cell lines over time. Moreover the bone marrow samples collected from a myeloma patient before and after a 12-month interval also revealed 2 new LOH loci (Figure 3C) of 1400 loci examined. These observations were additionally confirmed in 4 additional patients by collecting myeloma cells at 6-month intervals. These observations provide evidence of recombination or gene rearrangement events occurring in vivo. Each LOH “event” requires initial heterozygosity, a mutation such as gene conversion or deletion to eliminate either allele in a single originating cell, and then cell selection or population drift to reduce the frequency of the second allele to below detectable limits. Multiple LOH sites observed on different chromosomes (Figure 2C) argue against the possibility that they could have arisen from a single originating mutant cell already present in the population before the first observation. We further evaluated whether inhibition or induction of HR can affect the frequency of acquisition of new genetic changes in myeloma cells. Our data show that the inhibition of HR by siRNA directed at HsRAD51 significantly reduces the acquisition of new LOH, whereas its induction by nickel chloride significantly increases the number of new LOH sites in myeloma cells.
Genomic instability and continued acquisition of additional genetic changes are widely believed to underlie progression of cancer, including the development of more aggressive phenotypes as well as the development of drug resistance1,17,21,23–25; however, evidence of causation has been elusive. We therefore evaluated the effect of HR induction, and the consequent increase in genetic aberrations, on development of drug resistance in multiple myeloma. Induction of HR in myeloma cells cultured in the presence of a cytotoxic dose of dexamethasone (10−8 mol/L), led to development of dexamethasone resistance in a short period of time. These data recapitulate the observed in vivo development of drug resistance in patients after therapy and strongly implicate HR in this phenomenon. We therefore propose that continued acquisition of new genetic changes mediated by HR, as demonstrated here, leads to a progressive growth advantage to malignant cells, progression to increasingly malignant phenotypes, and development of drug resistance.
The main source of elevated/dysregulated HR activity in myeloma can be the deregulated/mutated recombinase and/or other HR protein(s). We chose hsRAD51 recombinase as a target to inhibit HR because it possesses multiple activities required for the process in a single protein and plays a predominant central role in this activity. However, the myeloma cells treated with RAD51-siRNAs had reduced expression of several other HR-related proteins, including Rad23B (implicated in damage detection), endonuclease G (implicated in initiation of HR), endonuclease GL2, endonuclease nTH1, endonuclease FEN1, RAD50, and RAD51 isoforms RAD51C and XRCC2 (Table 1). Reduced expression of these proteins may also have contributed to loss of HR activity in these cells.
Table 1.
Silencing of HsRAD51 recombinase leads to decreased expression of several homologous recombination genes
| Sr. no. | GenBank accession no. | Gene name | Fold reduction in gene expression |
|---|---|---|---|
| 1 | NM_002 874.1 | RAD23 homolog B (S cerevisiae) | 11.5 |
| 2 | NM_885728.1 | Endonuclease G-like 2 (ENDOGL2) | 4.3 |
| 3 | AF035B2D.1 | BRCA1-associated protein | 3.7 |
| 4 | U79718.1 | nth endonuclease Ill-like 1 (E coli) | 2.2 |
| 5 | NMJ04111.3 | Flap structure–specific endonuclease 1 | 1.6 |
| 6 | NM_885732.1 | RAD58 homolog | 1.4 |
| 7 | NMJ0287B.1 | RAD51 homolog C | 1.4 |
| 8 | NM_884435.1 | Endonuclease G | 1.4 |
| 9 | NMJ05431.1 | XRCC2 | 1.3 |
Gene expression profile was measured in ARP myeloma cells after transfection with control siRNA or siRNA directed at HsRAD51 and RAD51B as described in “Methods.” Fold reduction in the expression of HR genes in cells transfected with RAD51 siRNA relative to control cells is shown.
HR indicates homologous recombination; and siRNA, small inhibitory RNA.
We therefore propose that continued acquisition of new genetic changes mediated by HR, as demonstrated here, leads to a progressive growth advantage to malignant cells, progression to increasingly malignant phenotypes, and the development of drug resistance. Therefore, HR may not only be an important target for cancer therapy with a goal of inhibiting the development of invasive, metastatic or drug-resistant phenotypes, but also may be important in preventing progression of premalignant cells to overt malignancy.
Acknowledgments
This work was supported in part by grants from National Institutes of Health (Bethesda, MD; 1RO1CA125711-01A2), the Multiple Myeloma Research Foundation (Norwalk, CT) and NIH-P50-100007 Developmental Research Award to M.A.S. and from the Department of Veterans Affairs (Washington, DC) Merit Review Awards and NIH grants RO1-124929, P50-100007, and PO1-78 378 to N.C.M. The work also was supported in part by grants from the Department of Veterans Affairs (Merit Review and Research Career Scientist Award) to R.J.S.R.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.A.S. made the experimental plan, designed siRNAs, carried out recombination assays and gene expression studies, analyzed the data, and prepared the manuscript; R.J.S.R. assisted in data interpretation and analysis, and preparation of the manuscript; H.K. carried out cell culture studies, participated in siRNA transfections, and helped in the analysis and interpretation of data; R.B.B. carried out immunocytochemistry analyses; C.L. assisted in statistical analysis of gene expression data; and N.C.M. envisioned the study, and participated in its design and coordination, and also helped to draft the manuscript. All authors have read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nikhil C. Munshi, MD, Dana-Farber Cancer Institute, 44 Binney Street, M1B28, Boston, MA 02115; e-mail: Nikhil_Munshi@DFCI.Harvard.edu.
References
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 2.Shmookler Reis RJ, Goldstein S. Loss of reiterated DNA sequences during serial passage of human diploid fibroblasts. Cell. 1980;21:739–749. doi: 10.1016/0092-8674(80)90437-7. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava A, Norris JS, Shmookler Reis RJ, Goldstein S. c-Ha-ras-1 proto-oncogene amplification and overexpression during the limited replicative life span of normal human fibroblasts. J Biol Chem. 1985;260:6404–6409. [PubMed] [Google Scholar]
- 4.Tlsty TD. Normal diploid human and rodent cells lack a detectable frequency of gene amplification. Proc Natl Acad Sci U S A. 1990;87:3132–3136. doi: 10.1073/pnas.87.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sweezy MA, Fishel R. Multiple pathways leading to genomic instability and tumorigenesis. Ann N Y Acad Sci. 1994;726:165–177. doi: 10.1111/j.1749-6632.1994.tb52810.x. [DOI] [PubMed] [Google Scholar]
- 6.Roca AI, Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 7.Resnick MA, Skaanild M, Nilsson-Tillgren T. Lack of DNA homology in a pair of divergent chromosomes greatly sensitizes them to loss by DNA damage. Proc Natl Acad Sci U S A. 1989;86:2276–2280. doi: 10.1073/pnas.86.7.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker BS, Carpenter AT, Esposito MS, Esposito RE, Sandler L. The genetic control of meiosis. Annu Rev Genet. 1976;10:53–134. doi: 10.1146/annurev.ge.10.120176.000413. [DOI] [PubMed] [Google Scholar]
- 9.Cheng RZ, Shammas MA, Li J, Shmookler Reis RJ. Expression of SV40 large T antigen stimulates reversion of a chromosomal gene duplication in human cells. Exp Cell Res. 1997;234:300–312. doi: 10.1006/excr.1997.3649. [DOI] [PubMed] [Google Scholar]
- 10.Galli A, Schiestl RH. On the mechanism of UV and gamma-ray-induced intrachromosomal recombination in yeast cells synchronized in different stages of the cell cycle. Mol Gen Genet. 1995;248:301–310. doi: 10.1007/BF02191597. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Ayyadevera R, Shmookler Reis RJ. Carcinogens stimulate intrachromosomal homologous recombination at an endogenous locus in human diploid fibroblasts. Mutat Res. 1997;385:173–193. doi: 10.1016/s0921-8777(97)00054-2. [DOI] [PubMed] [Google Scholar]
- 12.Radman M, Jeggo P, Wagner R. Chromosomal rearrangement and carcinogenesis. Mutat Res. 1982;98:249–264. doi: 10.1016/0165-1110(82)90035-5. [DOI] [PubMed] [Google Scholar]
- 13.Rodarte-Ramón US. Radiation-induced recombination in Saccharomyces: the genetic control of recombination in mitosis and meiosis. Radiat Res. 1972;49:148–154. [PubMed] [Google Scholar]
- 14.Sengstag C. The role of mitotic recombination in carcinogenesis. Critical Reviews in Toxicology. 1994;24:323–353. doi: 10.3109/10408449409017922. [DOI] [PubMed] [Google Scholar]
- 15.Bishop JM. The molecular genetics of cancer. Science. 1987;235:305–311. [Google Scholar]
- 16.Honma M, Zhang LS, Hayashi M, et al. Illegitimate recombination leading to allelic loss and unbalanced translocation in p53-mutated human lymphoblastoid cells. Mol Cell Biol. 1997;17:4774–4781. doi: 10.1128/mcb.17.8.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shammas MA, Xia SJ, Shmookler Reis RJ. Induction of duplication reversion in human fibroblasts, by wild-type and mutated SV40 T antigen, covaries with the ability to induce host DNA synthesis. Genetics. 1997;146:1417–1428. doi: 10.1093/genetics/146.4.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Windle B, Draper BW, Yin YX, O'Gorman S, Wahl GM. A central role for chromosome breakage in gene amplification, deletion formation, and amplicon integration. Genes Devel. 1991;5:160–174. doi: 10.1101/gad.5.2.160. [DOI] [PubMed] [Google Scholar]
- 19.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 20.Bergsagel PL, Chesi M, Nardini E, Brents LA, Kirby SL, Kuehl WM. Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma. Proc Natl Acad Sci U S A. 1996;93:13931–13936. doi: 10.1073/pnas.93.24.13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn GK, Kurz BW, Cheng RZ, Shmookler Reis RJ. Homologous plasmid recombination is elevated in immortally transformed cells. Mol Cell Biol. 1989;9:4009–4017. doi: 10.1128/mcb.9.9.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leder P, Battey J, Lenoir G, et al. Translocations among antibody genes in human cancer. Science. 1983;222:765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- 23.Stürzbecher HW, Donzelmann B, Henning W, Knippschild U, Buchhop S. p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 24.Thyagarajan B, McCormick-Graham M, Romero DP, Campbell C. Characterization of homologous DNA recombination activity in normal and immortal mammalian cells. Nucleic Acids Res. 1996;24:4084–4091. doi: 10.1093/nar/24.20.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia SJ, Shammas MA, Shmookler Reis RJ. Elevated recombination in immortal human cells is mediated by HsRAD51 recombinase. Mol Cell Biol. 1997;17:7151–7158. doi: 10.1128/mcb.17.12.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson T, Lenhoff S, Rylander L, et al. High frequencies of chromosomal aberrations in multiple myeloma and monoclonal gammopathy of undetermined significance in direct chromosome preparation. Br J Haematol. 2004;126:487–494. doi: 10.1111/j.1365-2141.2004.05060.x. [DOI] [PubMed] [Google Scholar]
- 27.Sawyer JR, Waldron JA, Jagannath S, Barlogie B. Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genet Cytogenet. 1995;82:41–49. doi: 10.1016/0165-4608(94)00284-i. [DOI] [PubMed] [Google Scholar]
- 28.Shaughnessy J, Tian E, Sawyer J, et al. High incidence of chromosome 13 deletion in multiple myeloma detected by multiprobe interphase FISH. Blood. 2000;96:1505–1511. [PubMed] [Google Scholar]
- 29.Shammas MA, Liu X, Gavory G, Raney KD, Balasubramanian S, Shmookler Reis RJ. Targeting the single-strand G-rich overhang of telomeres with PNA inhibits cell growth and induces apoptosis of human immortal cells. Exp Cell Res. 2004;295:204–214. doi: 10.1016/j.yexcr.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Shammas MA, Neri P, Koley H, et al. Specific killing of multiple myeloma cells by (-)-epigallocatechin-3-gallate extracted from green tea: biologic activity and therapeutic implications. Blood. 2006;108:2804–2810. doi: 10.1182/blood-2006-05-022814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shammas MA, Shmookler Reis RJ, Akiyama M, et al. Telomerase inhibition and cell growth arrest by G-quadruplex interactive agent in multiple myeloma. Mol Cancer Ther. 2003;2:825–833. [PubMed] [Google Scholar]
- 32.Shammas MA, Simmons CG, Corey DR, Reis RJ. Telomerase inhibition by peptide nucleic acids reverses ‘immortality’ of transformed human cells. Oncogene. 1999;18:6191–6200. doi: 10.1038/sj.onc.1203069. [DOI] [PubMed] [Google Scholar]
- 33.Davies FE, Dring AM, Li C, et al. Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood. 2003;102:4504–4511. doi: 10.1182/blood-2003-01-0016. [DOI] [PubMed] [Google Scholar]
- 34.Rubnitz J, Subramani S. Rapid assay for extrachromosomal homologous recombination in monkey cells. Mol Cell Biol. 1985;5:529–537. doi: 10.1128/mcb.5.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munshi NC, Hideshima T, Carrasco D, et al. Identification of genes modulated in multiple myeloma using genetically identical twin samples. Blood. 2004;103:1799–1806. doi: 10.1182/blood-2003-02-0402. [DOI] [PubMed] [Google Scholar]
- 36.Munshi NC, Hideshima T, Carrasco D, et al. Identification of genes modulated in multiple myeloma using genetically identical twin samples. Blood. 2003 doi: 10.1182/blood-2003-02-0402. [DOI] [PubMed] [Google Scholar]
- 37.Shammas MA, Koley H, Batchu RB, et al. Telomerase inhibition by siRNA causes senescence and apoptosis in Barrett's adenocarcinoma cells: mechanism and therapeutic potential. Mol Cancer. 2005;4:24. doi: 10.1186/1476-4598-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shammas MA, Koley H, Beer DG, Li C, Goyal RK, Munshi NC. Growth arrest, apoptosis, and telomere shortening of Barrett's-associated adenocarcinoma cells by a telomerase inhibitor. Gastroenterology. 2004;126:1337–1346. doi: 10.1053/j.gastro.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 39.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Hong F. Cluster-Rasch models for microarray gene expression data. Genome Biol. 2001:2. doi: 10.1186/gb-2001-2-8-research0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 42.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 43.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dumur CI, Dechsukhum C, Ware JL, et al. Genome-wide detection of LOH in prostate cancer using human SNP microarray technology. Genomics. 2003;81:260–269. doi: 10.1016/s0888-7543(03)00020-x. [DOI] [PubMed] [Google Scholar]
- 45.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 46.Schild D, Lio YC, Collins DW, Tsomondo T, Chen DJ. Evidence for simultaneous protein interactions between human Rad51 paralogs. J Biol Chem. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- 47.Solomon E, Borrow J, Goddard AD. Chromosome aberrations and cancer. Science. 1991;254:1153–1160. doi: 10.1126/science.1957167. [DOI] [PubMed] [Google Scholar]
- 48.Bärlund M, Monni O, Kononen J, et al. Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res. 2000;60:5340–5344. [PubMed] [Google Scholar]
- 49.Hansen LT, Lundin C, Helleday T, et al. DNA repair rate and etoposide (VP16) resistance of tumor cell subpopulations derived from a single human small cell lung cancer. Lung Cancer. 2003;40:157–164. doi: 10.1016/s0169-5002(03)00026-6. [DOI] [PubMed] [Google Scholar]
- 50.Maacke H, Jost K, Opitz S, et al. DNA repair and recombination factor Rad51 is over-expressed in human pancreatic adenocarcinoma. Oncogene. 2000;19:2791–2795. doi: 10.1038/sj.onc.1203578. [DOI] [PubMed] [Google Scholar]
- 51.Sturgis EM, Clayman GL, Guan Y, Guo Z, Wei Q. DNA repair in lymphoblastoid cell lines from patients with head and neck cancer. Arch Otolaryngol Head Neck Surg. 1999;125:185–190. doi: 10.1001/archotol.125.2.185. [DOI] [PubMed] [Google Scholar]
- 52.Qiao GB, Wu YL, Yang XN, et al. High-level expression of Rad51 is an independent prognostic marker of survival in non-small-cell lung cancer patients. Br J Cancer. 2005;93:137–143. doi: 10.1038/sj.bjc.6602665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cousineau I, Abaji C, Belmaaza A. BRCA1 Regulates RAD51 Function in response to DNA damage and suppresses spontaneous sister chromatid replication slippage: implications for sister chromatid cohesion, genome stability, and carcinogenesis. Cancer Res. 2005;65:11384–11391. doi: 10.1158/0008-5472.CAN-05-2156. [DOI] [PubMed] [Google Scholar]