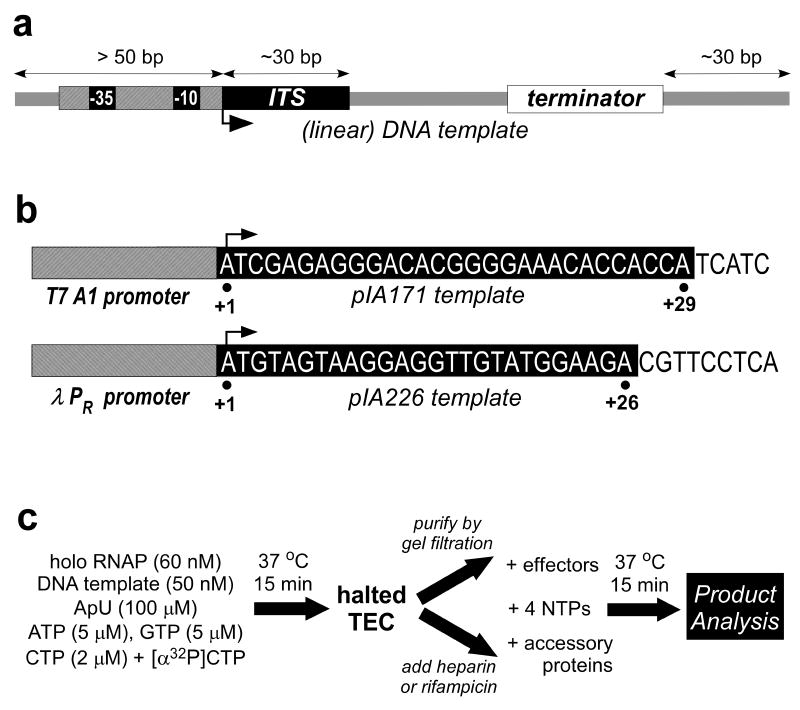
Fig. 1.
Single round termination assay design. (a) General features of the template. The template should encode a strong promoter (with -35 and -10 hexamers elements recognized by primary σ factors), a good initial transcribed sequence (ITS, at least 20 nt long and missing one of the bases), and a region of interest. For linear templates, at least 50 bp of DNA upstream of the transcription start site (a bent arrow, +1) and at least 30 bp downstream from the studied termination site(s) should be included to allow for proper recognition and for differentiation of terminated and readthrough products. (b) Examples of promoter-ITS combinations. The sequence of the non-template strand is shown, the RNA sequence is the same but with Us in place of Ts). pIA171 [32] has a phage T7A1 promoter followed by a 29 nt long T-less region; pIA226 [32] has a λ PR promoter and a 26 nt long C-less region. (c) Assay design (for pIA171). This protocol is suitable for RNAPs from E. coli or other mesophilic species; for RNAPs from thermophilic organisms (e.g., Thermus), the initiation reaction must be carried out at 55°C. Halted complexes will be radiolabeled in the 5′ end at several positions (10 if α-[32P]-GTP is used with pIA226-derived template).