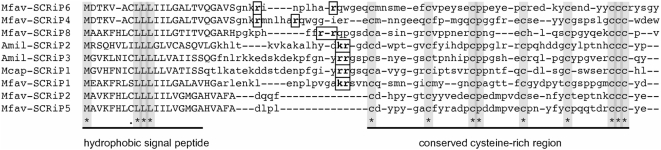
Figure 1. Multiple sequence alignments of SCRiPs identified in this study.
Predicted signal peptides are shown in upper case and conserved amino acid sites are indicated by an asterisk (100% similarity) or a dot (90% similarity) below the alignment. The N-terminal signal peptide region and C-terminal cysteine-rich domain are underlined. Boxed amino acids show potential proprotein convertase (PC) cleavage sites, which recognize the consensus motif [R/K]-[R/K], or [R/K]-(X)n-[R/K], where n = 2, 4 or 6.