Figure 1. mtDNA mutation load in colorectal and osteosarcoma cell lines.
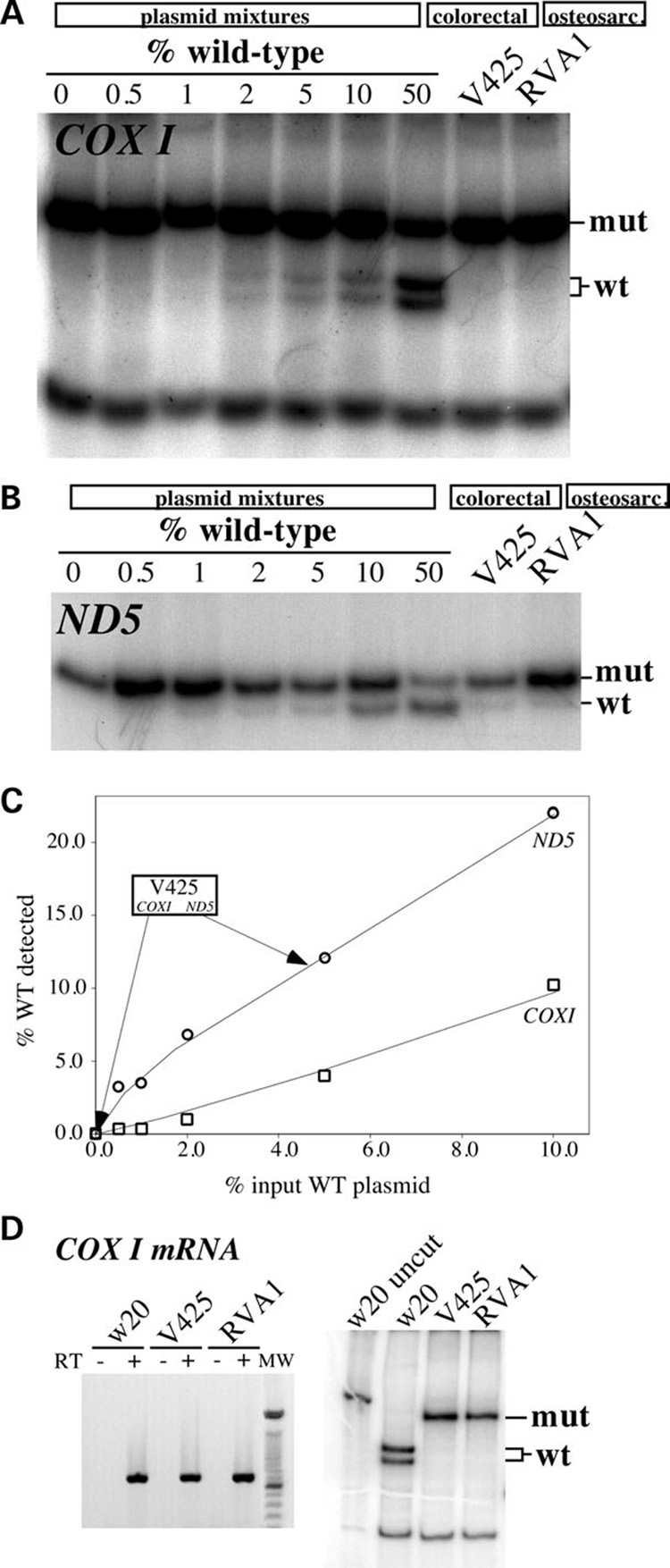
To assess the detection limits of the ‘last cycle hot’ PCR assay, plasmids containing mtDNA fragments harboring either the wild-type or mutated sequence were constructed for both the COXI and ND5 regions. Mutated and wild-type plasmids were mixed at known ratios and 10 ng of the sample was subjected to the ‘last cycle hot PCR’ followed by digestion with the appropriate restriction endonuclease (see Materials and Methods). (A) Analyses for the COXI G6264A mutation in COXI plasmid mixtures, the colorectal V425 and the osteosarcoma cybrid (RVA1). (B) A similar analysis for the 12418insA mutation in ND5 plasmid mixtures and the same cell lines. (C) Standard curve depicting the correlation between percentage input wild-type COXI- and ND5-containing plasmids and the percentage wild-type signal detected in the ‘last cycle hot PCR’/RFLP results. (D) Amplification (left) and ‘last cycle hot PCR’ analysis followed by restriction endonuclease digestion (right) of COXI mRNA amplified by RT-PCR.
