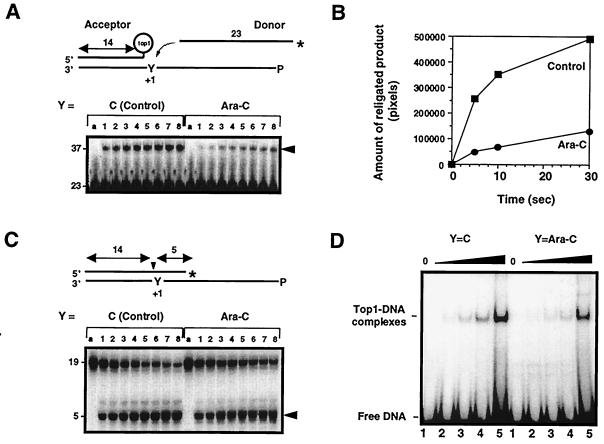
Figure 2.
Ara-C incorporation at the +1 position of a top1 cleavage site inhibits the religation of top1 cleavage complexes. (A) Ara-C inhibits the religation of top1 cleavage complexes. Acceptors were generated from substrates shown in C (see text for details). The unlabeled acceptors were then incubated with 10-fold excess of 3′-labeled, 23-mer donor strand for 5 s, 10 s, 0.5 min, 1 min, 3 min, 5 min, 10 min, or 30 min (lanes 1–8, respectively), and reactions were stopped with 0.5% SDS. (B) Quantitation of the gel shown in A. (C) Kinetics of top1-induced DNA cleavage. Oligonucleotides containing either C or ara-C at the +1 position were incubated with top1 for 5 s, 10 s, 0.5 min, 1 min, 3 min, 5 min, 10 min, or 30 min (lanes 1–8, respectively), and reactions were stopped with 0.5% SDS. (D) Effects of Ara-C incorporation on noncovalent top1 binding to DNA. Oligonucleotides with C or ara-C at the +1 position of the nonscissile strand (see Fig. 1B) were incubated with 0, 25, 50, 100, or 250 ng of purified human top1Y727Fp for 5 min at 25°C (lanes 1–5, respectively), and electromobility shift assay was performed. (A and C) Lanes a represent the DNA alone.