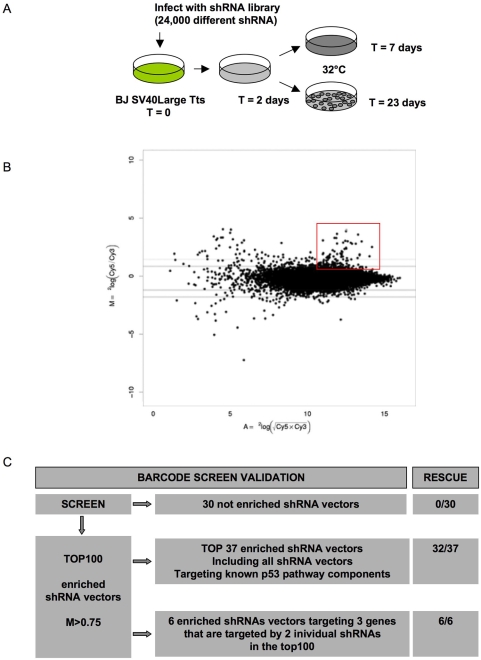
Figure 1. shRNA barcode screen identifies mediators of the p53 dependent cell cycle arrest.
a) Schematic outline of the BJtsLT genetic screen. BJtsLT cells were infected with the NKI shRNA library and were either left at 32°C or shifted to 39°C. After 7 days the cells at 32°C had reached confluency and were harvested. Cells at 39°C were harvested after 23 days after which they had formed visible colonies. b) Analysis of the relative abundance of shRNAs recovered from the BJtsLT barcode experiment. Data are normalized and plotted as M, the 2log (ratio Cy5/Cy3), versus A (2log(√intensity Cy3×Cy5)). The data are the average of two independent hybridization experiments performed in duplicate with reversed colour. A red box is drawn around the top 100 enriched shRNAs at 39°C. c) Schematic overview of selection criteria used to select hits from the shRNA barcode screen for further validation.