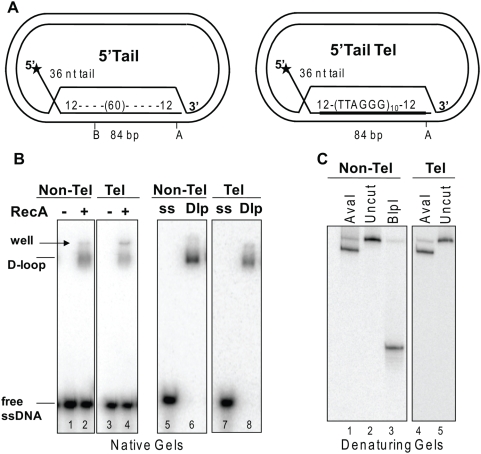
Figure 1. Construction of mobile plasmid D-loops.
A. Schematic of 5′ Tail non-telomeric and telomeric (Tel) plasmid D-loops. The star denotes the 5′ end radiolabel. The invading strand of the 5′Tail Tel D-loop base pairs with the plasmid to form an 84 bp duplex with ten (TTAGGG) repeats flanked by 12 bp of unique sequence. The telomeric repeats are scrambled in the non-telomeric D-loop. Restriction enzyme sites are indicated as A, AvaI; B, BlpI. B. RecA generated plamid D-loops are stable. Plasmids (300 µM nucleotides) were mixed with the 5′end-labeled complementary oligonucleotide (3.6 µM nucleotides) and RecA protein (4 µM) (lanes 2 and 4). D-loops (Dlp) were then purified from unincorporated oligonucleotide (ss) (lanes 6 and 8). Reactions were run on a 4–20% polyacrylamide native gel. C. The D-loop invading strand is fully base paired with the plasmid strand. Reactions (20 µl) contained 50 pM purified 5′Tail non-telomeric (Non-Tel) or telomeric (Tel) D-loops and 10 units of the indicated restriction enzyme. Products were run on a 14% polyacrylamide denaturing gel.