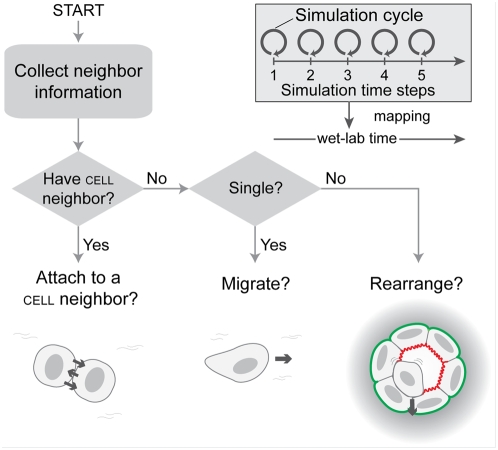
Figure 3. Encoded AT II cell step logic and the decision process.
Simulation time advances in steps corresponding to simulation cycles. Each simulation cycle maps to an identical interval of wet-lab time. During a simulation cycle, every culture component is given an opportunity to update. Every cell, selected randomly, executes a step function each simulation cycle to decide what action to take based on its internal state (clustered or single) and the composition of its adjacent neighborhood. Enabled cell actions are cell-cell attachment, cell migration, and rearrangement within a cluster. With a specified probability, a cell in either state will attach to one of its neighboring cells. A single cell can migrate to an adjacent space. A cell within a cluster can rearrange with other cells composing the cluster. Cell and clusters are specified parametrically to migrate randomly, chemotactically, along a cell density gradient, or using a combination of random and directional movement.