Figure 2.
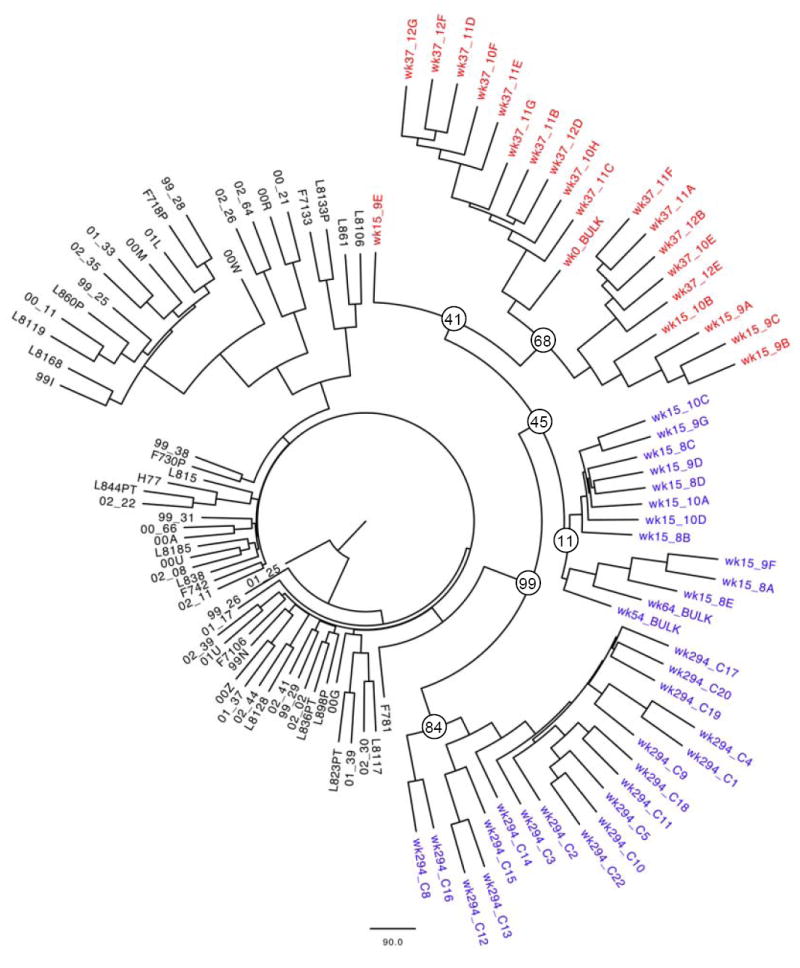
Bootstrap consensus tree of 100 replicates of maximum likelihood trees. The tree was constructed in PHYLIP (version 3.67, J. Felsenstein, University of Washington, available at http://evolution.genetics.washington.edu/phylip.html) based on 70 partial bulk NS3 sequences (nt 3331-4226 as aligned to H77) from the local Boston cohort and clonal sequences from the studied patient after removal of codon 155. While viral evolution over time is evident by somewhat distant clonal populations between weeks 0 and 294, all of the patient’s sequences (wild-type in blue and R155K variants in red) form a distinct cluster separate from sequences from all other patients, thus arguing against superinfection with a second viral strain during the six year observation period. The quasispecies in the mixed population at week 15 grouped in two distinct clusters: Variants harboring the R155K substitution grouped together with other variants from week 37 that also carried the R155K substitution, whereas variants at week 15 harboring the wild-type residue formed a separate phylogenetic group. These findings are suggestive of the coexistence of two phylogenetically distinct subpopulations in the quasispecies during the observation period. The bootstrap values of the corresponding nodes are indicated.
