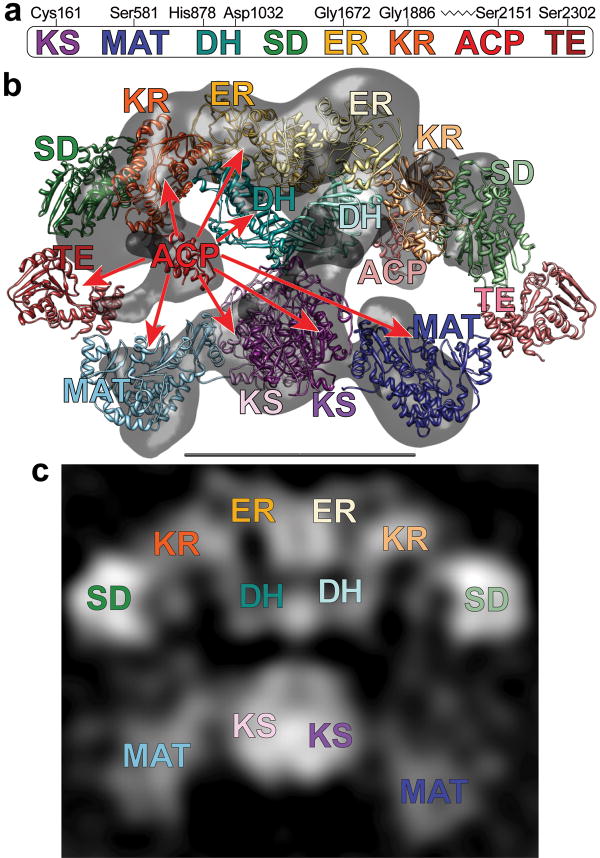
Figure 2.
(a) The domains of FAS are linearly arranged along the FAS polypeptide. Key active-site residues for the KS, MAT, DH and TE domains, the location of the glycine-rich motifs of the nucleotide-binding sites in the ER and KR domains, and the site of posttranslational phosphopantetheinylation in the ACP domain are marked (rat FAS numbering). (b) Crystal and cryo-EM structures capture different conformations of FAS. Atomic structures of individual catalytic domains were positioned according to the intermediate-resolution crystal structure7. ACP domains36 were fitted into remaining densities located below the KR domains in the cryo-EM structure (gray)6. Densities corresponding to the TE domains were not apparent in either the crystal structure or the cryo-EM structure but the domains are positioned near the outer edge of the two reaction chambers based on evidence presented in this article. Subunits in the FAS homodimer are depicted in a crossed-over arrangement with the domains of one subunit in faded colors. Catalytic contacts made by the ACP of one subunit are indicated by arrows. Flexibility of the KR-ACP linker and mobility of the phosphopantetheine are insufficient to explain contacts with the distant KS and MAT active sites. (c) A 2D class average calculated from images of FAS molecules preserved in stain has recognizable structural elements and shows good correspondence with the X-ray and cryo-EM 3D structures. The scale bar represents 100 Å.