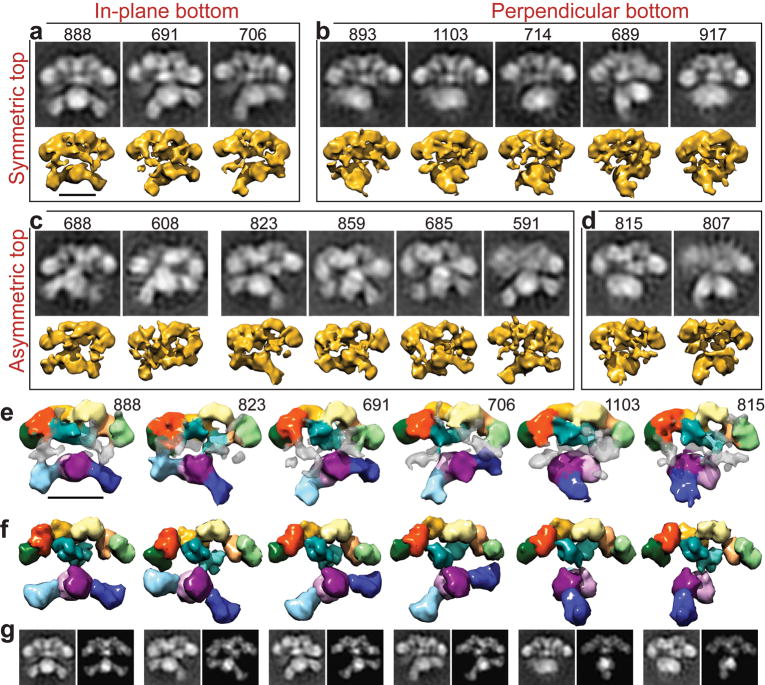
Figure 3.
Conformational variability of the Δ22-FAS in the absence of substrates. (a-d) Single particle images were classified (black and white panels) and corresponding 3D structures calculated (yellow). The number of particles in each class is indicated above its 2D class average. The domain arrangements in the upper portion of the structure range from predominantly symmetric (a,b) to strongly asymmetric (c,d). The lower domains are arranged with respect to the upper domains either in parallel, swinging from right to left (a,c), or swiveling about the narrow “waist” into a perpendicular arrangement (b,d). (e) 3D structures of the Δ22-FAS mutant were colored as in Figure 2 to indicate the regions that could be fitted with structures of the KS, MAT, DH, ER, KR, and SD. Regions of density that were not fitted (transparent grey) may accommodate the TE and/or ACP domains. (f) Atomic structures of individual domains were fitted into several RCT structures and filtered to match the resolution of the EM structures. (g) 2D projections of these fitted atomic structures (right image in each pair) closely resemble the 2D class averages (left image in each pair, also in a-d) that correspond to each of the 3D RCT reconstructions (directly above each pair in e). Scale bars represent 100 Å.