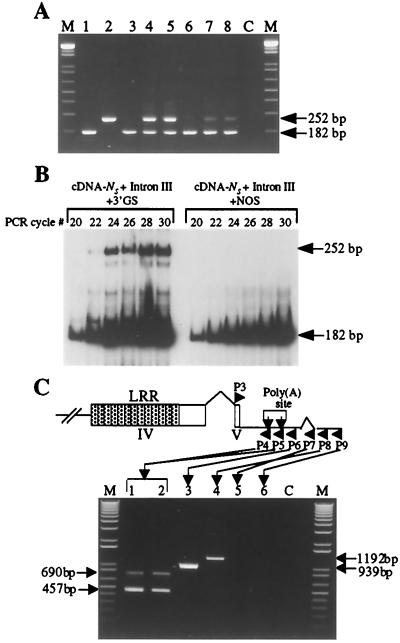
Figure 4.
The N gene 3′ GS is transcribed and influences alternative splicing. (A) Ethidium bromide-stained agarose gel showing RT-PCR products amplified by using primers P1 and P2 flanking intron III sequence. The first-strand cDNA was generated from poly(A)+ RNA extracted from plants expressing cDNA-NS:3′ GS (lane 1), cDNA-NL:3′ GS (lane 2), AE deletion (lane 3), cDNA-NS:3′ GS∷cDNA-NL:3′ GS (lanes 4 and 5), cDNA-NS + intron III (AE):NOS (lane 6), cDNA-NS + intron III (AE):3′ GS (lane 7), and SR1∷NN (lane 8) plants. Lane C is a negative control (for DNA contamination) in which poly(A)+ RNA without RT was used as a template. Lane M, 2 μg of 123-bp DNA ladder from GIBCO/BRL as molecular weight standard. (B) Autoradiograph of a polyacrylamide gel, showing radiolabeled RT-PCR products from the two N messages, NS and NL, generated by using radio-labeled P1 and unlabeled P2 primers that flank intron III. The first-strand cDNA was generated from poly(A)+ RNA extracted from cDNA-NS + intron III (AE):3′ GS (Left) or cDNA-NS + intron III (AE):NOS (Right) plants. Each lane represents products generated during the exponential phase of amplification (20–30 cycles). (C) Ethidium bromide-stained agarose gel showing 3′ rapid amplification of cDNA ends (RACE) PCR products amplified by using primer P3 that anneals to exon V and primers P4-P9 that anneal to sequences downstream of the N gene stop codon. The first-strand cDNA was generated from nuclear RNA extracted from N gene-containing plants and used as templates for 3′ RACE. Lane C is a negative control (for DNA contamination) in which poly(A)+ RNA without RT was used as a template. Lane M, 2 μg of 123-bp DNA ladder from GIBCO/BRL as molecular weight standard. Arrows to the right and left of the gel indicate sizes in bp of 3′ RACE products.