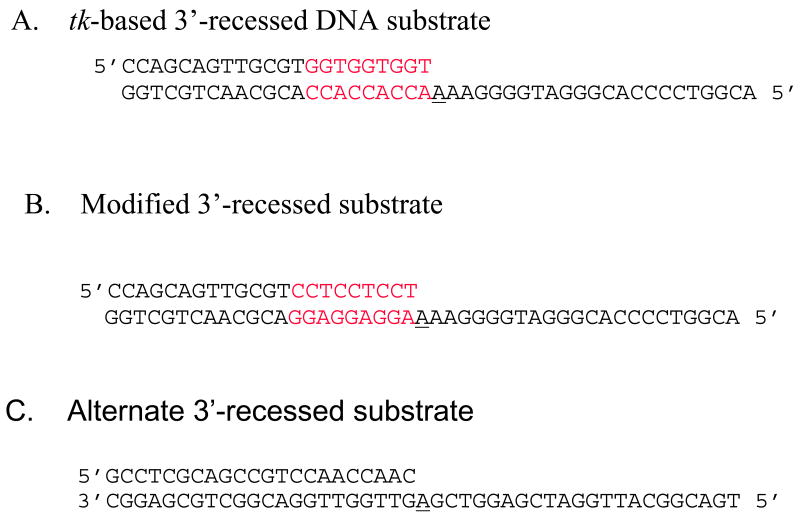
Figure 2. DNA substrates used in kinetics experiments.
A) Original HSV-tk-based 3′-recessed substrate. This DNA substrate mimics position 243 of the HSV-tk gene and the surrounding sequence. B) Modified 3′-recessed DNA substrate. In this DNA substrate the primer and template sequences adjacent to the templating base have been flipped in order to determine if a misalignment-mediated slippage mechanism leads to misincorporation. C) Primer/template unrelated to the HSV-tk gene. In each case, the template is 45 nucleotides long and primer is 22 nucleotides long. The primer and template of the DNA duplex were phosphorylated with [γ-32P]ATP at the 5′-end using T4 polynucleotide kinase. The templating base is underlined and the red bases denote the three-nucleotide repeats.