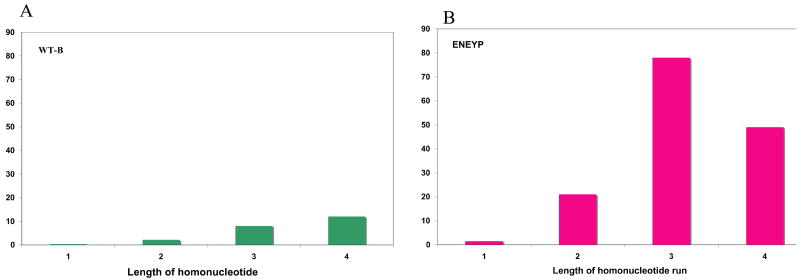
Figure 4. Frameshift frequency as a function of the homonucleotide run length.
A) WT and B) ENEYP variant. Frameshift frequencies at each repeat length (1 is a non-iterated base) were adjusted for number of potential sites of template slippage and total number of bases in the tk target sequence. For example, there are seventeen independent occurrences of 1-base frameshifts at two nucleotide repeats out of a total fifty-one 1-base frameshift. Seventeen was divided by fifty-one to obtain the proportion of 1-base frameshifts and the value was multiplied by the frameshift frequency. The resultant value was then divided by the number of potential sites for frameshifts at two-nucleotide repeats. The X-axis represents the length of the nucleotide repeats and the y-axis represents the frameshift frequency.