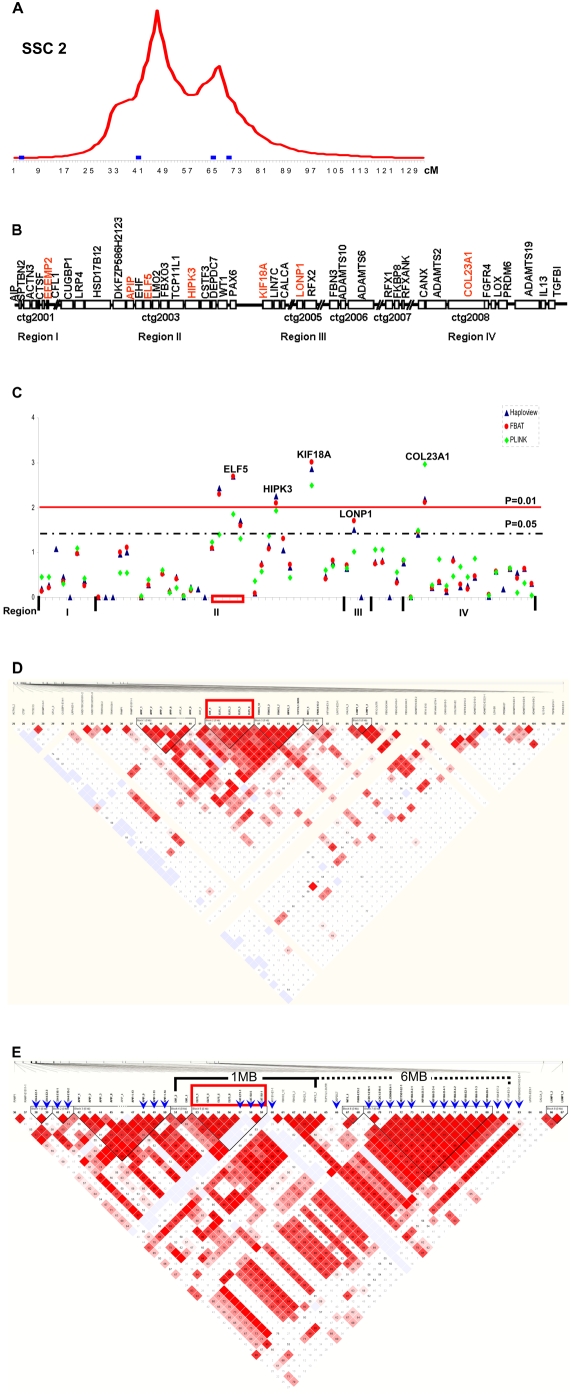
Figure 1. Extensive positional candidate gene analyses of four porcine genomic regions for hernia development on SSC2.
A, Hernia QTL on SSC2. Genomic region around 48 cM was significantly linked to risk of hernia development. Three regions (I, at 3 cM; II at 42 cM; III, at 66 cM) and a fourth region (IV at 70 cM) could be potentially linked to scrotal hernia. These regions were indicated by the blue frames. B, The gene map was built based on the latest porcine physical FPC (fingerprint contigs) map (Sanger Porcine Genome Sequencing Project) (Table S1). Genes in contigs 1, 3, 5 and 8 on SSC2 were selected to cover the four regions, respectively. Contigs 6 and 7 were also used to search for candidate genes. C, Single marker association analyses. SNPs of genes located in region II were significantly associated with pig hernia in Pietrain-based line. The small red box includes SNPs in ELF5 significantly associated with scrotal hernia. D, LD heatmap was constructed by Haploview. Genes in region II were found to be in high LD with each other. The red box indicates the common haplotype formed by ELF5-1, ELF5-3, ELF5-5, ELF5-8 and EHF1 was significantly associated with scrotal hernia. E, 26 new SNP markers were added into the region II region (blue arrowhead), with focus on the 1 MB region including EHF-ELF5-CAT, and the 6 MB region covering KIF18A. Re-sequencing work on KIF18A did not detect any further association signal. The red box defines the haplotype composed of ELF5-1, ELF5-3, ELF5-5, ELF5-8, CAT-E11-1, CAT-5U-5 and CAT-5U-1 to be highly significantly associated with scrotal hernia.