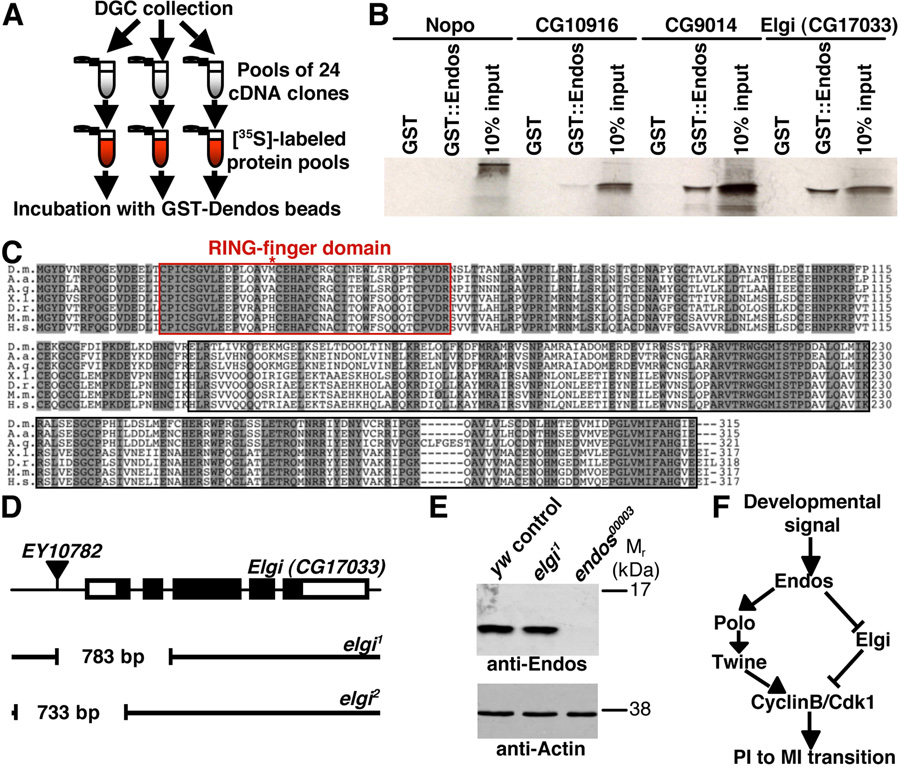
Figure 4. Endos binds to a putative E3 ubiquitin ligase encoded by elgi.
(A) DIVEC screen used to identify Endos interactors. (B) Autoradiogram showing that the GST::Endos fusion protein specifically binds to the closely related CG9014 and Elgi E3 ubiquitin ligases but not to more distant E3s, such as Nopo and CG10916. (C) Elgi shares 81%, 80%, 57%, 57%, 56%, and 54% amino acid identity with the A. aegypti, A. gambiae, X. laevis, D. rerio, M. musculus, and H. sapiens homologs, respectively. Red box marks RING domain. Shaded amino acid residues represent identities. Asterisk indicates the first methionine for predicted Elgi2 protein. (D) elgi alleles generated by imprecise excision of EY10782. Thick bars represent elgi exons (coding region in black). Gaps in black lines indicate deleted regions in elgi1 and elgi2. (E) Western showing normal Endos levels in elgi1 ovaries. Actin used as loading control. (F) Model for the role of Endos in oocyte meiotic maturation. Endos controls Polo kinase and, perhaps indirectly, Twine levels, leading to activation of CyclinB/Cdk1. By binding and inhibiting Elgi, Endos may have a parallel role in refining the timing of meiotic maturation.