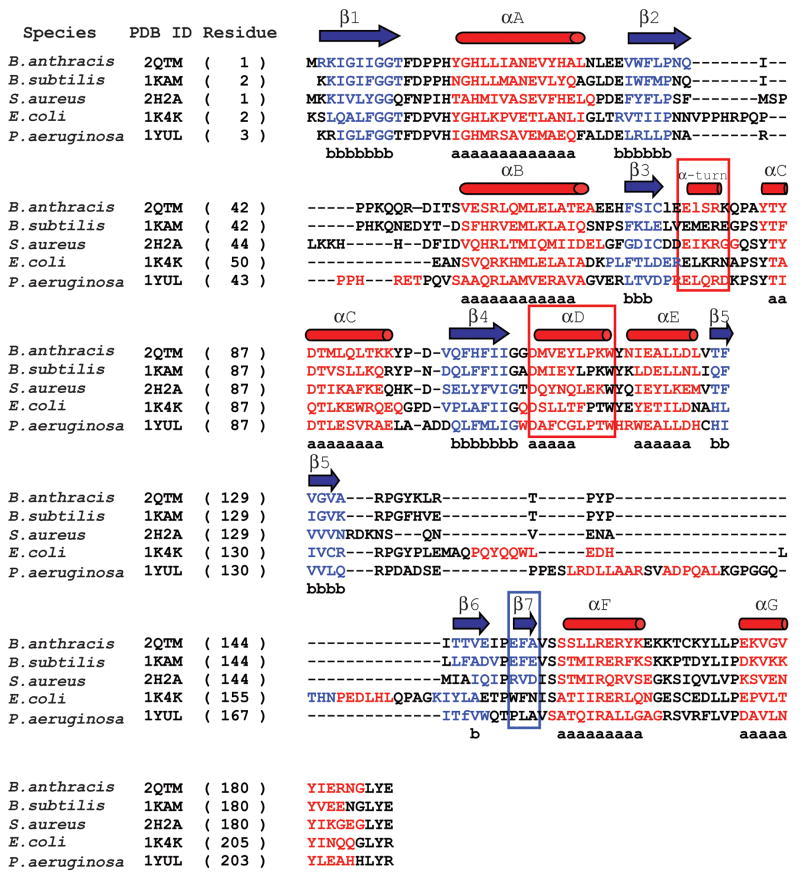
Figure 6. Structure-based sequence alignment of bacterial NMAT’s.
A structure-based sequence alignment of the B. anthracis NMAT with B. subtilis NMAT, S. aureus NMAT, E. coli NMAT, and P. aeruginosa NMAT. B. anthracis NMAT secondary structure is shown above the alignment, where blue arrows and red cylinders denote β-sheets and α-helices, respectively. Residues involved in β-sheet and α-helix formation are colored blue and red, respectively. Black colored residues represent loop regions. Lower case “b’s” represent conserved β-sheet formation and lower case “a’s” represent conserved α-helix formation. The red boxes represent the secondary structural elements that are different between B. anthracis and B. subtilis NMAT structures. The blue box indicates the different secondary structural elements between gram (−) and gram (+) bacteria. This figure was generated using STRAP (http://www.charite.de/bioinf/strap/) and the secondary structure of B. anthracis NMAT was manually added. STRAP inserted many unnecessary gaps and therefore for clarity the gaps were removed in the region of the α-helix D.