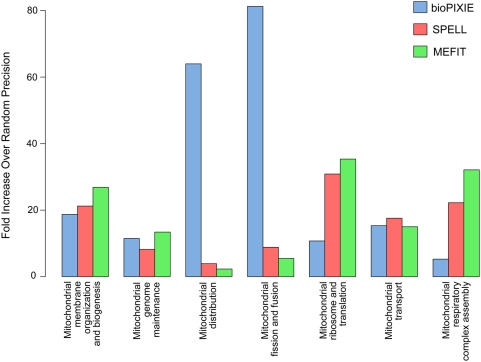
Figure 5. Biological differences between the three computational prediction methods.
We evaluated which aspects of mitochondrial biology were targeted by each computational function prediction method. Even though all three methods learned and were evaluated using the same set of training genes, the methods differ in the sub-groups of mitochondrial biology on which they focused. SPELL and MEFIT are both based solely on gene expression microarray data, which explains their strong coverage of the mitochondrial ribosome and translation sub-group. bioPIXIE is based on diverse data, including physical binding data, which explains its strong coverage of sub-groups involving mitochondrial motility and physical interactions.