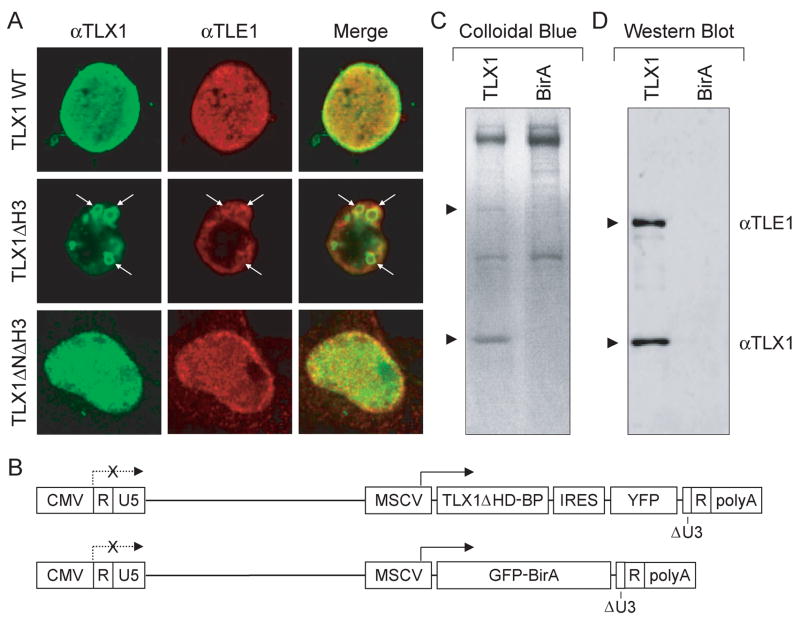
Fig. 2.
TLX1 interacts with TLE1 in vivo. (A) 293T cells transiently expressing TLX1 and TLE1 (indicated to the left of the panels) were labeled with anti-TLX1 (green) and anti-TLE1 (red) antibodies, and immunofluorescent staining was analyzed by confocal laser scanning microscopy. Overlapping regions of protein distribution appear yellow. Ring-like structures are indicated by white arrows. (B) Schematic representation of the self-inactivating lentiviral vectors used to produce biotinylated TLX1. Abbreviations: CMV, cytomegalovirus immediate early enhancer-promoter; R, direct repeat; U5, unique 5′ region of long terminal repeat; MSCV, murine stem cell virus promoter; TLX1ΔHD-BP, TLX1 homeodomain deletion mutant with COOH-terminal biotinylation peptide tag; IRES, internal ribosome entry site; YFP, yellow fluorescent protein; GFP-BirA, green fluorescent protein-BirA biotin ligase fusion protein; ΔU3, deleted unique 3′ region of long terminal repeat; polyA, polyadenylation signal. (C) SDS-PAGE gel stained with Colloidal blue after streptavidin pulldown of extracts from SupT1 cells expressing the TLX1ΔHD-BP protein (TLX1) or the GFP-BirA protein alone. Arrows point to the positions of TLX1 and TLE1 (identified by mass spectrometry). (D) Western blot analysis of the same material described in (C).