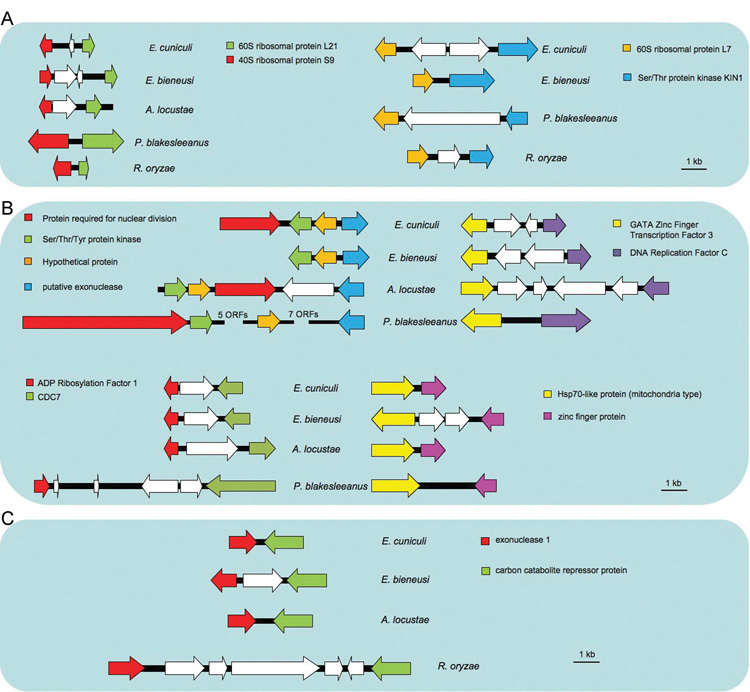
Figure 2. Examples of conserved synteny and relaxed synteny found in zygomycetes and microsporidia.
(A) Conserved synteny among P. blakesleeanus, R. oryzae, and the microsporidia. Syntenic 60S (L21) and 40S (S9) protein genes are conserved throughout fungi (see Supplementary Fig. 6). Another synteny of ribosomal protein and kinase genes is observed in two zygomycete genomes and two microsporidian genomes (E. cuniculi and E. bieneusi).
(B) Conserved synteny between P. blakesleeanus and the three microsporidia.
(C) Conserved synteny between R. oryzae and the three microsporidia. White arrows indicate additional non-syntenic genes.
With the exception of two ribosomal proteins (L21 and S9), none of the syntenic gene clusters shown in panels (A), (B) and (C) were conserved in other ascomycete (A. nidulans), basidiomycete (C. neoformans), or chytridiomycete (B. dendrobatidis) fungi. The genomes of P. blakesleeanus and R. oryzae were compared to the E. cuniculi genome using BLASTp. Homologs in E. bieneusi and A. locustae were identified using tBLASTn.