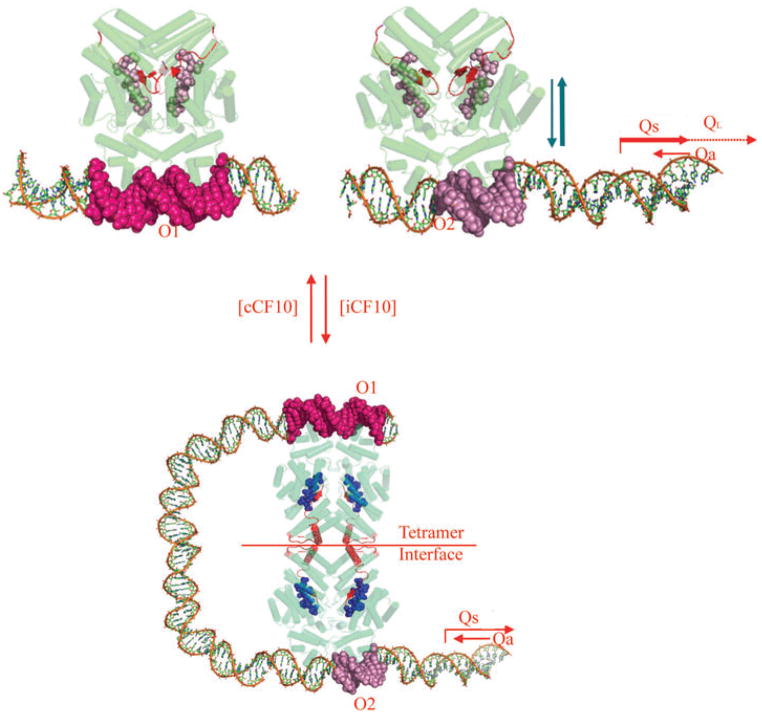
Fig. 1.
PrgX regulation of the prgQ conjugative transfer operon of plasmid pCF10. Predicted functional activity of intracellular PrgX bound to cCF10 or iCF10 are shown in the top and bottom panels respectively. PrgX binds two operator sites in pCF10 DNA. The O2 site overlaps the prgQ promoter and PrgX occupancy of this site inhibits prgQ transcription, but the binding interaction at this site is very weak unless the O1 site is also occupied.
Top. PrgX is shown in green with barrels indicating α-helices and the C-terminal region in red. Pheromone cCF10 is illustrated by red spheres. Target DNA is shown in multicolour. Pheromone binding alters the C-terminal region of PrgX such that the interaction of pairs of PrgX dimers to form tetramers is weakened. Pheromone-bound PrgX ineffectively represses PQ. The resulting high levels of the Qs transcript produced under these conditions titrates all of the Qa RNA produced from the complementary DNA strand, allowing QL production (right).
Bottom. The data presented in this paper indicate that PrgX/iCF10 complexes are comprised of the inhibitor peptide ligand (blue spheres) bound to the same PrgX cleft as cCF10, but that differences in interaction of the bound peptide with the C-terminus of PrgX actually stabilize a tetramer interface that is predicted to cause formation of a DNA loop in vivo. This loop increases occupancy of the operator sites, leading to reduced expression from PQ. The resulting lower level of Qs RNA allows it to be completely bound by the constitutively expressed Qa, resulting in termination of prgQ transcription. Even in the uninduced state, repression of PQ is not complete, and a significant basal level of Qs, which encodes iCF10, is produced.