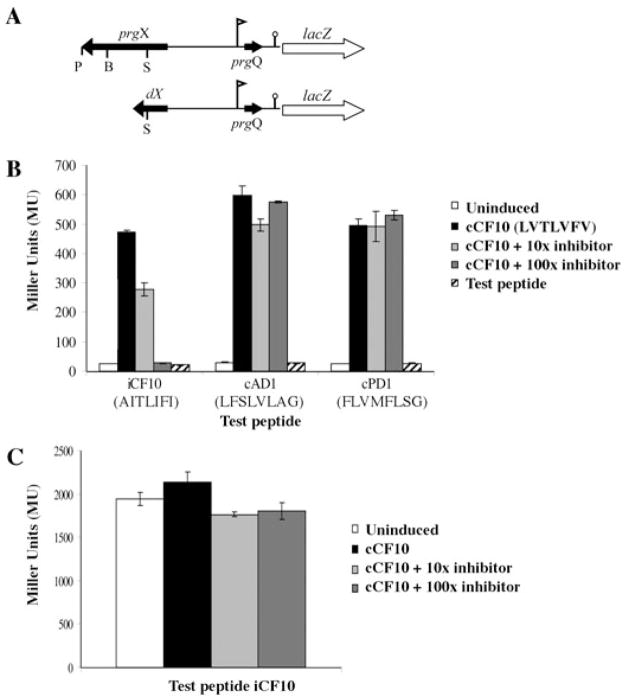
Fig. 2. Inhibition of pheromone-inducible transcription from the prgQ promoter by iCF10 requires PrgX.
A. Schematic drawing of pheromone-inducible p043lacZ reporter construct (top) and the p043dX derivative lacking a functional prgX gene (bottom). Flag indicates the direction and location of PQ, lollipop indicates IRS1 sequence where prgQ transcription terminates in uninduced cells, genes are indicated by arrows showing approximate size and direction of transcription. P = PstI, B = BglII, S = SpeI restriction enzyme cleavage sites.
B. Effects of addition of various combinations of cCF10 and other peptides on prgQ expression in OG1RF(p043lacZ). Cells were exposed to various combinations of cCF10 and either iCF10, cAD1 or cPD1 at 10-fold and 100-fold molar excess. iCF10, cAD1 and cPD1 were also tested in the absence of cCF10 addition for their ability to act as inducers in this system; the amino acid sequence of each peptide is shown. cCF10 = cCF10 at 5 ng ml−1, 10X inhibitor = cCF10 at 5 ng ml−1 + test peptide at 50 ng ml−1, 100X inhibitor = cCF10 at 5 ng ml−1 + test peptide at 500 ng ml−1, Test peptide = test peptide at 25 ng ml−1, no cCF10.
C. Effects of various combinations of iCF10 and cCF10 on prgQ transcription in OG1RF containing p043lacZ versus p043dX. Same amounts of cCF10 and iCF10 as in (B); cAD1 and cPD1 not used as test peptides in this experiment.