Table 3.
S4 regions of voltage-gated K+ and H+ channels and a voltage-sensing phosphatase (VSP)
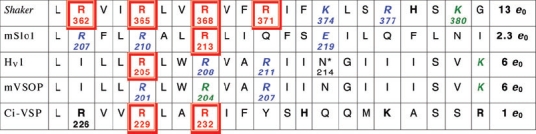 |
A sequence of 21 amino acids in the putative S4 regions of the Shaker K+ channel, the maxi-K calcium-activated K+ channel (mSlo), the human proton channel (HV1), the mouse proton channel (mVSOP), and a voltage-sensing phosphatase in Ciona (Ci-VSP). Potentially charged amino acids are indicated in bold. Single neutralization mutations at the numbered positions in red boxes reduce the gating charge; those in italics (in blue) do not affect gating charge, or were inconclusive (in green) (Aggarwal & MacKinnon, 1996; Seoh et al. 1996; Bezanilla, 2000; Murata et al. 2005; Ramsey et al. 2006; Sasaki et al. 2006; Ma et al. 2006; Musset et al. 2008a; Hossain et al. 2008). Total gating charge (e0) estimates are listed in the final column; for HV1 and mVSOP, these are from the limiting slope method (Musset et al. 2008a). The mouse R204Q mutant did not express well; R207Q was identical to wild type, and for R201Q activation was faster and the gH–V relationship was shifted by −50 mV and had a slightly steeper slope (z = 1.9 versus 1.4 for wt) (Sasaki et al. 2006). In human proton channels, gating was faster for all three Arg mutants, and for R205A the midpoint was shifted positively, and the slope (from gH–V relationships) was less steep by 1/3 (zδ= 0.57 versus 0.90 for wt) (Ramsey et al. 2006). The VSD of the Shaker K+ channel can be transformed into a proton channel by the R362H mutation (Starace & Bezanilla, 2004), and R362X where X = Cys, Ala, Ser or Val produces non-selective cationic ‘omega’ current through the voltage sensor (Tombola et al. 2005). *The N214R mutation greatly attenuates conduction (Tombola et al. 2008). A ClustalW alignment of proteins containing S4 regions homologous to HV1 was manually adjusted to reflect predicted transmembrane regions. In some cases, plausible alternative alignments can be obtained by shifting a sequence by three residues. These previously unpublished alignments were generously provided by S. M. E. Smith (Emory University).
