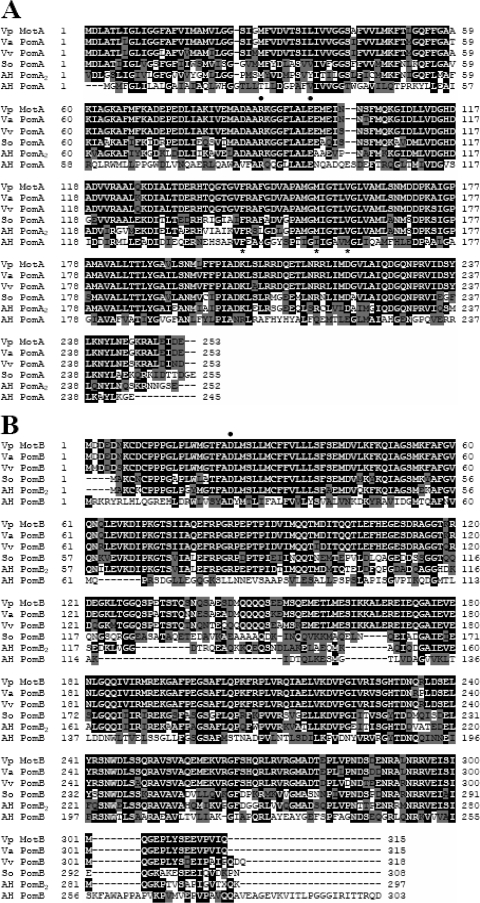
FIG. 5.
(A) Sequence alignment of a Vibrio parahaemolyticus PomA homologue (Vp MotA), Vibrio alginolyticus PomA (Va PomA), Vibrio vulnificus PomA (Vv PomA), Shewanella oneidensis PomA (So PomA), A. hydrophila AH-3 PomA2 (AH PomA2), and PomA (AH PomA). (B) Sequence alignment of a V. parahaemolyticus PomB homologue (Vp MotB), V. alginolyticus PomB (Va PomB), V. vulnificus PomB (Vv PomB), S. oneidensis PomB (So PomB), and A. hydrophila AH-3 PomB2 (AH PomB2) and PomB (AH PomB). The deduced amino acid sequences were aligned using Clustal W. White letters with a black background indicate identical amino acid residues, and black letters with a gray background indicate similar amino acid residues. The black circles over the residues show conserved charged residues in proton and sodium motors, and the asterisks show charged residues essential in Vibrio sodium-dependent motor function.