FIG. 1.
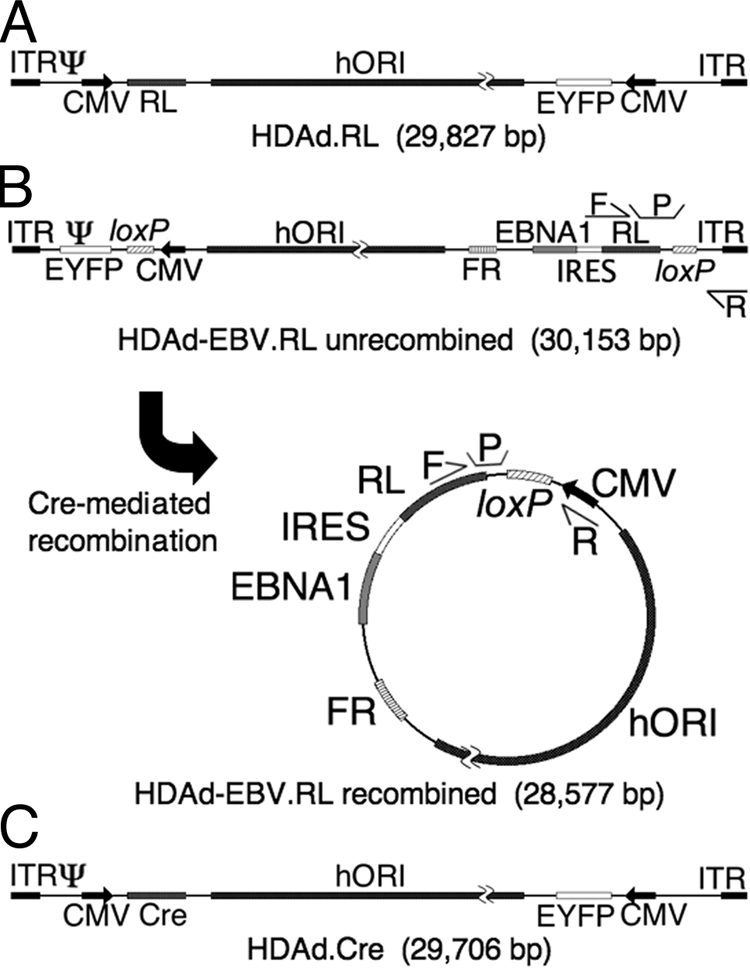
Structure of HDAd vectors. Three vectors were used in the course of these experiments. Each contains an EYFP gene expressed from a constitutive human CMV major immediate-early promoter (CMV); a 19-kb region of human genomic DNA from chromosome 10 for use as a stuffer and as an origin of replication (hORI) (21); and two regions from adenovirus, the inverted terminal repeats (ITR) that function as replication origins and the viral packaging signal (Ψ) that are required in cis for vector packaging and amplification. (A) HDAd.RL is a traditional HDAd vector that was used as a control in these experiments. It expresses RL constitutively from a CMV promoter. (B) HDAd-EBV.RL is a hybrid vector that is designed to deliver a circular EBV episome into target cells by means of a linear HDAd genome. In its linear form, the EBV episome elements are flanked by parallel recognition sites for Cre recombinase (loxP). In the presence of Cre recombinase, the intervening region is excised and circularized as shown. This places a CMV promoter upstream of an expression cassette that contains the RL transgene, an encephalomyocarditis virus internal ribosomal entry site (IRES), and the EBNA1 gene. The episome also caries the FR region that functions with EBNA1 protein to provide a maintenance function to the episome in replicating cells. The relative location of two sets of qPCR primers (F and R) and probes (P) are indicated. One set flanks one of the loxP sites in the linear conformation of the hybrid vector, and a second set flanks the loxP site in the circularized conformation. These were used in qPCR experiments to determine the relative fraction of recombined and unrecombined vector. (C) HDAd.Cre is a traditional HDAd vector that is designed to express Cre recombinase in target cells by means of a CMV promoter. Coinfection of a target cell by both HDAd-EBV.RL and HDAd.Cre is required to form the circular EBV episome.
