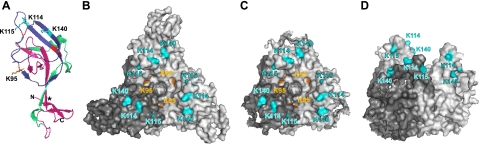
FIG. 7.
Model of 19-kDa GP1 and locations of key receptor binding residues. (A) Ribbon diagram of 19-kDa GP1 showing key recombinant RBRs used in this study, including RBR-7 (residues 90 to 149; dark blue), RBR-1 (residues 57 to 149; dark blue and green), and RBR-12 (residues 33 to 193; dark blue, green, and pink). The side chains of K95, K114, K115, and K140 are shown. Cysteines involved in disulfide bonds are shown in red. An asterisk (*) denotes a contact point between the β1 and β13 strands. (B) Surface rendering (top view) of the GP1,2Δ trimer structure. One monomer is colored dark gray for clarity. (C and D) Model of GP1,2Δ trimer after cleavage by cathepsins B and L, viewed from the top (C) and side (80° rotation) (D). Residues that decreased RBR-1-Fc binding when replaced with Ala are shown in all figures (K95, orange; K114, K115, and K140, cyan). The graphic representations were based on PDB file 3CSY (14) and rendered with Pymol. Note that the depictions in panels A, C, and D are only models that assume no conformational changes in GP1 following proteolytic priming.