Abstract
Herpes B virus (BV) naturally infects macaque monkeys and is a close relative of herpes simplex virus. BV can zoonotically infect humans to cause a rapidly ascending encephalitis with ∼80% mortality. Therefore, BV is a serious danger to those who come into contact with these monkeys or their tissues and cells. MicroRNAs are regulators of gene expression, and there have been reports of virus-encoded microRNAs. We hypothesize that BV-encoded microRNAs are important for the regulation of viral and cellular genes. Herein, we report the discovery of three herpes B virus-encoded microRNAs.
Cercopithecine herpesvirus 1 (also called herpes B virus, herpesvirus simiae, or B virus [BV]) is an alphaherpesvirus that naturally infects macaque monkeys. In macaques, the virus typically causes a self-limiting disease similar to herpes simplex virus (HSV) disease in humans (27). In striking contrast, BV infection of humans has resulted in the death of ∼80% of untreated individuals (27). Even with timely antiviral therapy, 20% of those infected die (10). For these reasons, the Centers for Disease Control and Prevention recommend that BV be propagated only in biosafety level 4 (BSL-4) laboratories. Additionally, the virus and viral DNA are designated select agents by the U.S. Department of Justice. The E2490 strain of BV has been completely sequenced (18), and the organization of the BV genome is almost identical to that of the HSV genome. The highly pathogenic nature of BV and its prevalence in macaque monkeys, which are commonly used in research, make BV a serious concern to animal handlers. Understanding the basis of BV pathogenesis will help lead to improved safety for those who may become exposed to BV. Moreover, considering that HSV is the most frequent cause of aseptic encephalitis (13), furthering our understanding of the biology of BV—an extreme example of a herpesvirus that causes encephalitis—may provide important insights into HSV encephalitis.
Over the last few years, microRNAs (miRNAs) have emerged as important regulators of gene expression (1, 3). Most miRNAs are 21 to 23 nucleotides (nt) long and are derived from longer primary miRNAs. The regulation of gene expression by miRNAs depends on the degree of complementarity between the miRNA and its target sequence and the location of the target sequence in the regulated mRNA (reviewed in references 1 and 3). Imperfect complementarity to a sequence in the 3′ untranslated region (3′ UTR) usually results in the inhibition of translation of the target mRNA, while perfect complementarity to a region in the coding sequence usually results in the cleavage of the target mRNA. miRNAs have been shown to be encoded by several DNA viruses (12), including viruses belonging to each of the alpha-, beta-, and gammaherpesvirus subfamilies (6-8, 11, 14, 15, 19, 21, 23, 25, 26). The regulation of cellular transcripts by virally encoded miRNAs has been reported previously (22). Additionally, examples of virally encoded miRNAs that regulate viral genes transcribed from the opposite strand of the genome have been described elsewhere (4, 24-26). It has been proposed that herpesviruses employ miRNAs to modulate the expression of their own genes, including genes in the immediate early kinetic class, as part of their strategy to enter the host and maintain latency (17, 26). We hypothesize that BV-encoded miRNAs are important in viral pathogenesis. As a first step, we have identified BV-encoded miRNAs by using a combination of computational methods and Northern blot hybridizations.
Computational prediction of BV-encoded miRNAs.
To computationally predict BV-encoded miRNAs, we used an algorithm previously developed to predict HSV-encoded miRNAs (11) and the published sequence of BV strain E2490 (GenBank accession no. NC_004812) (18). The only modification from the method used for HSV type 1 miRNA prediction was that the maximum permissible G+C content within the 21-nt query sequences was raised from 70 to 80%. This adjustment was made to account for the higher G+C content of the BV genome than of the HSV genome (74% in BV versus 68% in HSV type 1). To confirm the effectiveness of the algorithm, it was run against the Epstein-Barr virus (EBV) genome. All experimentally verified EBV miRNAs were successfully identified (G. Li and X.-J. Wang, unpublished data). When the algorithm was run against the BV genome, 17 genomic loci that contained 19 putative miRNA precursors were identified (Li and Wang, unpublished). Three of the predicted miRNAs (Fig. 1) were experimentally validated; these miRNAs are discussed below.
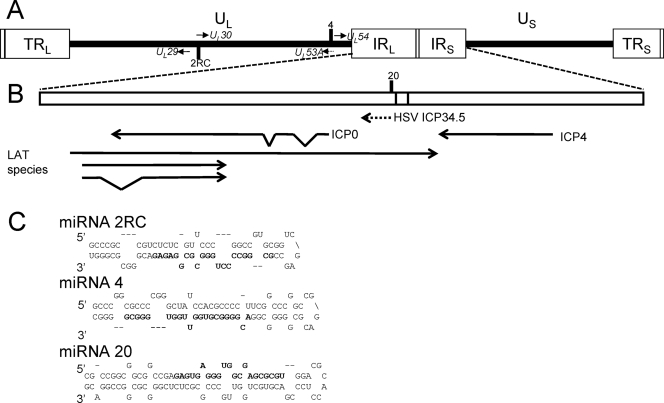
FIG. 1.
Genomic locations of the identified BV-encoded miRNAs. (A) Diagram of the prototypic arrangement of the BV genome, with the unique long (UL) and unique short (US) sequences (thick lines) bracketed by terminal repeat (TR) and internal repeat (IR) regions (boxes). The locations of the predicted BV-encoded miRNA precursors are marked by a vertical line and the miRNA number. A number above the horizontal line indicates that the miRNA is transcribed from left to right. A number below the horizontal line indicates that the miRNA is transcribed from right to left. The genes closest to each miRNA are noted in italics. (B) Expanded diagram of the internal repeat region noting the location of BV miRNA 20. The relative sizes, locations, and orientations of the transcripts encoded by the corresponding region of HSV are shown (ICP34.5 is marked with a dotted line as it is known to be absent in BV). The transcriptional map of HSV is shown, as this region of BV has yet to be carefully mapped. Bent lines indicate sequences removed by splicing. LAT, latency-associated transcript. (C) Primary sequences of the predicted pre-miRNAs. The predicted sequences of the processed mature miRNAs are shown in bold.
Validation of computationally predicted miRNAs by Northern blot hybridization.
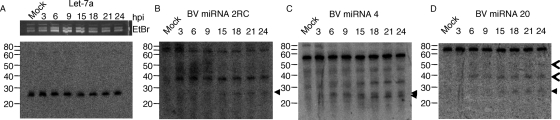
Northern blot hybridization of RNA isolated from BV-infected cells was used to experimentally validate the predicted miRNAs. In the BSL-4 laboratory at the Southwest Foundation for Biomedical Research, San Antonio, TX, African green monkey (Vero) cells were infected with BV strain E2490 at a multiplicity of infection of 10. At various times postinfection, cells were scraped from the dishes and pelleted, the supernatant was removed, and the pellet was flash frozen in liquid nitrogen and then stored at −80°C until it was processed. RNA was harvested from infected cells by using the mirVana miRNA isolation kit according to the instructions of the manufacturer (Ambion, Austin, TX). RNA species of <200 nt recovered with the mirVana kit, alongside 32P-labeled Decade markers (Ambion), were electrophoretically resolved on denaturing 15% polyacrylamide gels containing 8 M urea in Tris-borate-EDTA (2). The gels were then submerged in 0.5 μg/ml ethidium bromide in Tris-borate-EDTA for 3 min to permit the visualization of the RNA on a UV transilluminator. RNA was transferred onto a nylon membrane (BrightStar-Plus; Ambion) by using a semidry apparatus (Bio-Rad, Hercules, CA) and then cross-linked by using a Stratalinker (Stratagene, Cedar Creek, TX). The membranes were probed with DNA oligonucleotides (Integrated DNA Technologies, Coralville, IA) end labeled with [32P]ATP (MP Biomedicals, Irvine, CA). Unincorporated nucleotides were removed using Micro Bio-Spin columns according to the instructions of the manufacturer (Bio-Rad). Only probes with specific activity exceeding 5 × 106 dpm/pmol, as measured using a liquid scintillation counter, were used. The sequences of the probes that successfully identified BV-encoded miRNAs are as follows: for miRNA 2RC, GCCCGTCTCTCCGCGCCCAGGGGCCGC; for miRNA 4, GCCCCGCCCACCAACCACGCCCCGT; for miRNA 20, CGTCCACGCGCTCGCCACCCTCACTC; and for cellular miRNA let-7a, ACTATACAACCTACTACCTCA. Hybridizations were performed in ULTRAhyb-Oligo buffer according to the instructions of the manufacturer (Ambion). Probed membranes were exposed to phosphor storage screens (GE Life Sciences, Pittsburgh, PA) and analyzed with a Storm phosphorimager (GE Life Sciences). Prior to reprobing, membranes were stripped by being soaked two times in boiling 0.1% sodium dodecyl sulfate solution for 15 min. Figure 2 shows the phosphor images of the membranes probed for BV miRNAs 2RC, 4, and 20. BV miRNAs 2RC and 20 appear to be expressed with late kinetics. For BV miRNA 4, there are two virus-specific bands of approximately the correct size for miRNAs; one is more abundant at early times, and the other is more abundant at late times.
FIG. 2.
Identification of BV-encoded miRNAs. RNA from mock-infected cells or from cells isolated at the indicated number of hours postinfection (hpi) was resolved on a denaturing acrylamide gel. (A) Northern blot hybridization loading controls. tRNA from the ethidium bromide (EtBr)-stained gel is shown in the top panel. Below is shown a phosphorimage of the membrane probed for let-7a. To the left are shown the positions of the RNA markers. (B to D) The data for each predicted miRNA that was verified by Northern blot hybridization are shown. Virus-specific miRNA species are marked with a closed arrowhead. Virus-specific RNA species larger than expected for miRNAs are marked with an open arrowhead.
Expression of miRNAs in cells transfected with plasmids carrying the miRNA sequences.
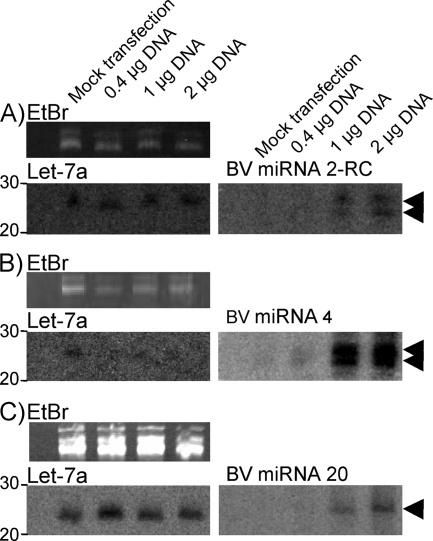
We addressed the possibility that the signals observed on the Northern blots represented breakdown products of viral or cellular transcripts that had occurred as a result of virus infection, rather than miRNAs. For example, in HSV infection, virion host shutoff protein (Vhs) degrades viral and cellular transcripts (20), and BV encodes a Vhs homolog (18). Each miRNA sequence, together with ∼200 nt of flanking sequence, was amplified from BV strain E2490 DNA by PCR and cloned into pCR2.1TOPO by using a TOPO-TA cloning system (Invitrogen). The PCR primers were as follows: miRNA 2RC forward primer, GATCCGTCGTTGCTAGGCAA, and reverse primer, GTTGTAAAAGTTCTCGCGCGG; miRNA 4 forward primer, TCCATCATCCTGTCCGGGATAGC, and reverse primer, CGTCCGCGTCCTCGAAGT; and miRNA 20 forward primer, TCGGGTCCGCGCTACCGGAG, and reverse primer, GCGCGCACTCGCGAGGGA. Plasmids were sequenced to determine the orientation of the insert and to ensure that PCR errors were not introduced, and the plasmids were then digested with BamHI and XhoI. BV sequences were subcloned into BamHI- and XhoI-digested pcDNA4/TO. Vero cells (8 × 105) were seeded onto a 60-mm dish and, after 16 h, transfected with various amounts of each plasmid by using Effectene (Qiagen). Small RNA was isolated 24 h posttransfection, and Northern blot hybridization was performed as detailed above. Increasing amounts of each miRNA, concomitant with the amounts of plasmid DNA introduced by transfection, were observed in transfected cells (Fig. 3). Two bands were observed for both miRNA 2RC and miRNA 4. In each case, there was a band that comigrated with that for let-7a and one that likely corresponds to an RNA species a single base smaller. Similarly, multiple species have been observed previously when other miRNAs have been examined by Northern blot hybridization (see, e.g., reference 14), and cloning studies have determined that the 3′ ends of miRNAs often vary by one or two nucleotides (16).
FIG. 3.
Detection of miRNAs from plasmid-expressed primary miRNAs. Each miRNA sequence, together with ∼200 nt of flanking sequence, was cloned into an expression vector. Vero cells were transfected with various amounts of each plasmid (DNA amounts are indicated by the lane labels above panel A), and the small RNA was harvested at 24 h posttransfection. Each panel is organized similarly. The top image shows tRNAs on the ethidium bromide (EtBr)-stained gel as a loading control. The image directly below shows the membrane probed for the cellular miRNA let-7a. To the right is shown the membrane probed for the indicated BV miRNA. Arrowheads mark miRNA species.
What are the potential targets of the BV-encoded miRNAs?
It is possible that these BV-encoded miRNAs regulate the expression of cellular genes by repressing translation through imperfect interactions in the 3′ UTR. More work will be required to elucidate these targets. However, in the light of recent reports demonstrating that virus-encoded miRNAs can regulate the expression of genes via perfect complementarity (4, 24-26), we have considered this scenario to be a possibility for the BV-encoded miRNAs.
BV miRNA 2 is antisense relative to a sequence ∼100 nt upstream of the initiating AUG of the UL30 gene, which encodes the catalytic subunit of the DNA polymerase. Therefore, miRNA 2 may regulate the expression of UL30 in the same way that the EBV-encoded miRNA miR-BART2 regulates the expression of the EBV DNA polymerase (4). Interestingly, the expression of HSV DNA polymerase is inhibited via a short sequence in the 5′ UTR of the UL30 gene; however, it is unclear whether an miRNA is involved (5).
BV miRNA 4 is antisense relative to a region in the UL53A open reading frame. The UL53A gene is a putative two-exon gene that was identified previously by statistical analysis of the BV genome (18). This locus contains one of the few substantive differences between the BV genome and the HSV genome; thus, it has the potential to be important for the differences in pathogenesis between BV and HSV in humans. If UL53A is determined to be generated in infected cells, it will be interesting to see if BV miRNA 4 is involved in regulating its expression and how this regulation affects pathogenesis.
BV miRNA 20 is antisense relative to a sequence that, in HSV, would encode ICP34.5 and be the only antisense target for this miRNA (20). ICP34.5 is an important HSV neurovirulence factor (9). Paradoxically, BV lacks a homolog of ICP34.5 (18). We have recently discovered BV-encoded transcripts that are antisense relative to BV miRNA 20 (M. Harden and A. Griffiths, unpublished observations); however, further work will be required to determine their functions and whether they are targets of BV miRNA 20.
It is worthy of note that none of the BV-encoded miRNAs have homologs in HSV. It has been noted previously that virus-encoded miRNAs tend not to be conserved, even between closely related viruses (12). A relevant exception comes from studies of EBV and the related rhesus lymphocryptovirus, which show that several miRNAs are conserved between the two viruses (8, 15, 19). There may be BV-encoded miRNAs that are homologous to HSV-encoded miRNAs that are yet to be discovered, and vice versa. However, it is interesting to speculate that the lack of homology between the currently reported miRNAs encoded by BV and HSV may be a factor in the outcome of BV infection of humans versus macaques. Herein, we report three BV-encoded miRNAs—the first miRNAs discovered to be encoded by a risk group 4 agent. There is accumulating evidence that miRNAs are important for the pathogenesis of herpesviruses; thus, these miRNAs and the genes they regulate may reveal important novel targets for antiviral therapy.
Acknowledgments
We thank Adriana Mejia and Nicole Henry for excellent technical assistance. We also thank Jean Patterson and Ricardo Carrion for help in the BSL-4 laboratory, Angela Pearson for helpful comments on the manuscript, and the members of the laboratories of S.-J. Gao and Ken Izumi for helpful comments and advice.
This work was supported by a grant from the Southwest Foundation Forum and was conducted in facilities constructed with support from Research Facilities Improvement Program grant number C06 RR012087 from the NCRR.
Footnotes
Published ahead of print on 14 January 2009.
REFERENCES
- 1.Ambros, V. 2004. The functions of animal microRNAs. Nature 431350-355. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seridman, J. A. Smith, and K. Struhl. 2007. Current protocols in molecular biology. John Wiley and Sons, Inc., Hoboken, NJ.
- 3.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. [DOI] [PubMed] [Google Scholar]
- 4.Barth, S., T. Pfuhl, A. Mamiani, C. Ehses, K. Roemer, E. Kremmer, C. Jaker, J. Hock, G. Meister, and F. A. Grasser. 2008. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 36666-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, K. F., and D. M. Coen. 2008. Inhibition of translation by a short element in the 5′ leader of the herpes simplex virus-1 DNA polymerase transcript. J. Virol. 8277-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnside, J., E. Bernberg, A. Anderson, C. Lu, B. C. Meyers, P. J. Green, N. Jain, G. Isaacs, and R. W. Morgan. 2006. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J. Virol. 808778-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 1025570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, X., A. Schafer, S. Lu, J. P. Bilello, R. C. Desrosiers, R. Edwards, N. Raab-Traub, and B. R. Cullen. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou, J., E. R. Kern, R. J. Whitley, and B. Roizman. 1990. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science 2501262-1266. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, J. I., D. S. Davenport, J. A. Stewart, S. Deitchman, J. K. Hilliard, and L. E. Chapman. 2002. Recommendations for prevention of and therapy for exposure to B virus (cercopithecine herpesvirus 1). Clin. Infect. Dis. 351191-1203. [DOI] [PubMed] [Google Scholar]
- 11.Cui, C., A. Griffiths, G. Li, L. M. Silva, M. F. Kramer, T. Gaasterland, X. J. Wang, and D. M. Coen. 2006. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J. Virol. 805499-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen, B. R. 2006. Viruses and microRNAs. Nat. Genet. 38(Suppl.)S25-S30. [DOI] [PubMed] [Google Scholar]
- 13.Davison, K. L., N. S. Crowcroft, M. E. Ramsay, D. W. Brown, and N. J. Andrews. 2003. Viral encephalitis in England, 1989-1998: what did we miss? Emerg. Infect. Dis. 9234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grey, F., A. Antoniewicz, E. Allen, J. Saugstad, A. McShea, J. C. Carrington, and J. Nelson. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 7912095-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundhoff, A., C. S. Sullivan, and D. Ganem. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12733-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim, L. P., N. C. Lau, E. G. Weinstein, A. Abdelhakim, S. Yekta, M. W. Rhoades, C. B. Burge, and D. P. Bartel. 2003. The microRNAs of Caenorhabditis elegans. Genes Dev. 17991-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy, E., J. Vanicek, H. Robins, T. Shenk, and A. J. Levine. 2008. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. USA 1055453-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perelygina, L., L. Zhu, H. Zurkuhlen, R. Mills, M. Borodovsky, and J. K. Hilliard. 2003. Complete sequence and comparative analysis of the genome of herpes B virus (Cercopithecine herpesvirus 1) from a rhesus monkey. J. Virol. 776167-6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304734-736. [DOI] [PubMed] [Google Scholar]
- 20.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. Herpes simplex viruses, p. 2502-2601. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 21.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 799301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samols, M. A., R. L. Skalsky, A. M. Maldonado, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 3e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schafer, A., X. Cai, J. P. Bilello, R. C. Desrosiers, and B. R. Cullen. 2007. Cloning and analysis of microRNAs encoded by the primate gamma-herpesvirus rhesus monkey rhadinovirus. Virology 36421-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan, C. S., and D. Ganem. 2005. MicroRNAs and viral infection. Mol. Cell 203-7. [DOI] [PubMed] [Google Scholar]
- 25.Tang, S., A. S. Bertke, A. Patel, K. Wang, J. I. Cohen, and P. R. Krause. 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc. Natl. Acad. Sci. USA 10510931-10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umbach, J. L., M. F. Kramer, I. Jurak, H. W. Karnowski, D. M. Coen, and B. R. Cullen. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454780-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitley, R. J., and J. Hilliard. 2007. Cercopithecine herpes virus 1 (B virus), p. 2889-2903. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]