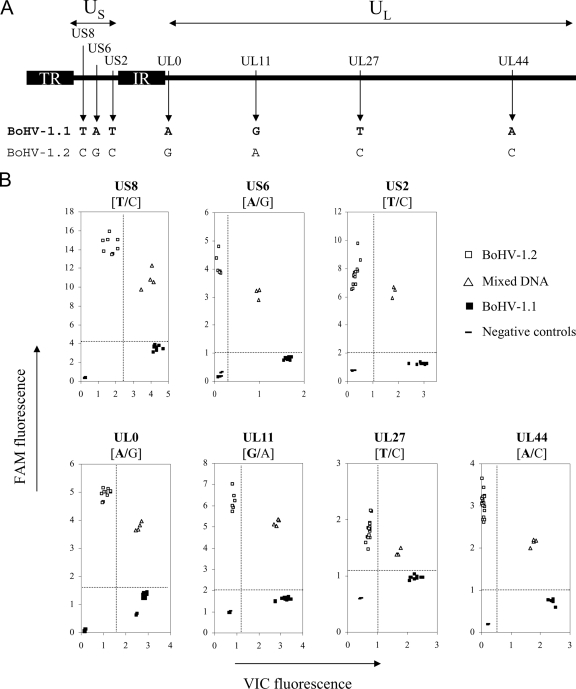
FIG. 2.
TaqMan genotyping assays discriminate seven SNPs differentiating BoHV-1.1 and BoHV-1.2 genomes. (A) Organization of the BoHV-1 genome including two unique sequences, a long one (UL) and a short one (US). The latter is flanked by two repeated and inverted sequences (IR, internal repeat; TR, terminal repeat). Shown are the localizations of the seven point mutations detected in seven BoHV-1 genes that are targeted by TaqMan genotyping assays. The nucleotides indicate the allelic version discriminating BoHV-1.1 (in boldface type) and BoHV-1.2 at each position. (B) Endpoint reading of the fluorescence generated during PCR amplification of seven target genes discriminating BoHV-1.1 (▪) and BoHV-1.2 (□). BoHV-1.1 DNA hybridized with the VIC-labeled probes, giving rise to an intense fluorescence along the x axis during amplification. BoHV-1.2 alleles hybridized with the FAM-labeled probes, resulting in a high fluorescence signal along the y axis. Different ratios of BoHV-1.1 and -1.2 DNA (1/10, 1/1, and 10/1) were mixed together and submitted to the analysis; these samples (▵) were hybridized with both the FAM- and VIC-labeled probes, resulting in increased fluorescence signals along both axes. Dot plot data were generated from endpoint readings of the fluorescence recorded after TaqMan PCR assays were performed on total DNA quantities ranging from 1 to 20 ng.