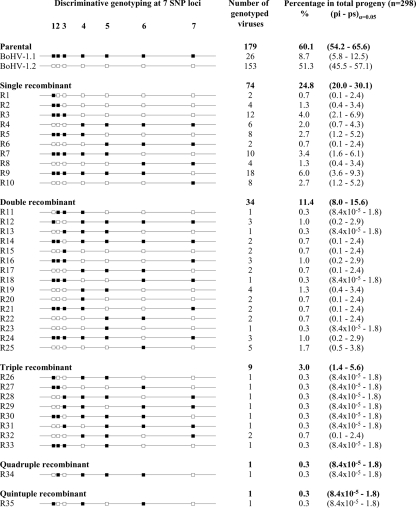
FIG. 3.
Characterization and division of the progeny virus genotypes issued from in vitro coinfection with BoHV-1.1 and BoHV-1.2. A total of 298 randomly plaque-purified virions issued from a BoHV-1.1 and BoHV-1.2 coinfection experiment were submitted to TaqMan genotyping assays at seven selected SNP sites in BoHV-1, the US8 (1), US6 (2), US2 (3), UL0 (4), UL11 (5), UL27 (6), and UL44 (7) genes. The allelic discrimination data are represented by small squares at each SNP position. Filled squares (▪) indicate the alleles inherited from BoHV-1.1; empty squares (□) indicate the alleles inherited from BoHV-1.2. Progeny virus genotypes were distributed in 37 different SNP combinations: 2 parental (P1 and P2) and 35 recombinants (R1 to R35). The configurations of recombinants were further classified as single, double, triple, quadruple, and quintuple recombinants by counting the recombination events present in the virus genotype. Quantitative data are presented as the absolute number of genotyped viruses classified in each configuration and as the relative frequency in the total progeny. The confidence intervals (α = 0.05) were calculated by determining the lower (pi) and upper (ps) limits of frequency in a binomial sampling distribution.