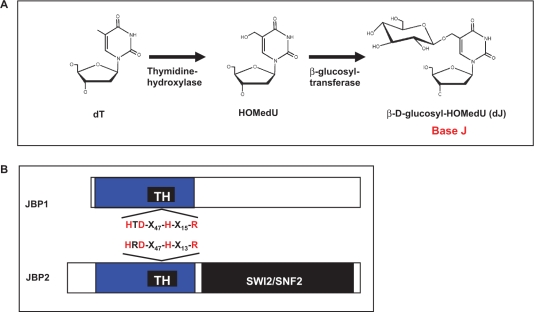
Figure 1.
Base J is synthesized in a two-step pathway. (A) Schematic model of the J-biosynthesis pathway. Step 1 involves the oxidation of thymidine (dT) residues in DNA forming the intermediate HOMedU. In step 2 the intermediate (HOMedU) is then glucosylated, by a glucosyl-transferase enzyme. This is then glucosylated, by a glucosyl-transferase enzyme, forming base J (dJ). The initial site-specific hydroxylation of thymidine, by TH, is thought to represent the key regulatory step of J biosynthesis. (B) Schematic diagram of the two proposed THs: JBP1 and JBP2. Shown in blue is the N-terminal region shared between both proteins. Within this region is the ∼70-amino-acid motif, indicated by the solid black box, which is related to the functional domain of members the Fe2+/2-oxoglutarate dependent hydroxylase family. Indicated above and below the putative TH motif (TH) for JBP2 and JBP1 is the amino acid signature shared among members of this hydroxylase family. The four key residues within this motif that have been implicated in catalysis by other members of this family of proteins (including JBP1), and which were mutated to alanine in JBP2 for the studies presented here, are highlighted in red. The C-terminal SWI2/SNF2 domain of JBP2 is shown in black.