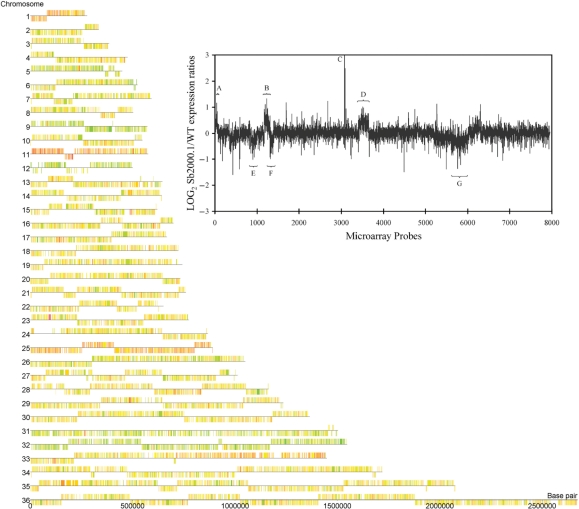
Figure 4.
Chromosome map of L. infantum Sb2000.1/WT gene expression modulation. DNA microarrays data were analyzed with the GeneSpring GX3.1 software to illustrate the Sb2000.1/WT expression ratios on a chromosome map of L. infantum. Orange to red features indicate genes overexpressed in Sb2000.1, whereas pale to bright green features indicate genes downregulated in Sb2000.1. Yellow features indicate genes equally expressed in both samples. (Insert) Log2-transformed Sb2000.1/WT expression ratios plotted as a function of the chromosomal location of every probes represented on the full-genome microarrays from chromosome 1 (left end) to chromosome 36 (right end). Probes are plotted by coordinates along each chromosome. Vertical bars represent the log2-transformed expression ratio of individual genes. Whereas most of the genes represented by the microarrays do not show a modulated expression, some chromosomes are entirely overexpressed or downregulated at the RNA level. (A) chromosome 1; (B) chromosome 11; (C) overexpressed locus on chromosome 23; (D) chromosome 25; (E) chromosome 9; (F) chromosome 12; (G) chromosome 32. The plot represents the average values of five independent hybridizations.