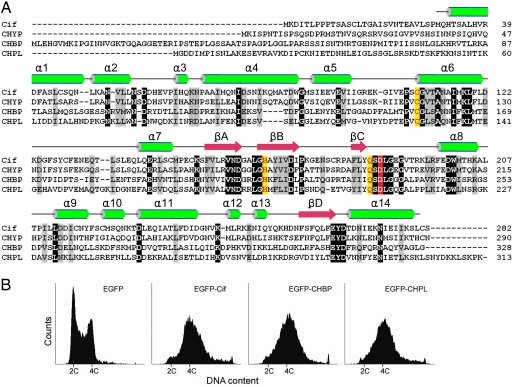
Fig. 1.
Identification of the Cif family of cell cycle modulators. (A) Multiple sequence alignment of the Cif family of effectors. The Cif family was identified by BLAST searches using the sequence of Cif from EPEC. Alignment was generated by using the ClustalW program with manual adjustments based on predicted secondary structures. The protein names are indicated at the beginning of the alignment: CHYP, from Y. pseudotuberculosis YPIII (ORF name, YPK_1971); CHPL, from P. luminescens TTO1 (ORF name, plu2515); CHBP, from B. pseudomallei K96243 (ORF name, BPSS1385). Secondary structure elements determined from the CHBP crystal (see Fig. 2) are indicated on top of the sequence with α-helix in green and β-strand in pink. Identical residues are highlighted in black, and similar residues are in gray. Functionally important residues (see Figs. 3B and 5) are colored in yellow or red. (B) Effects of the Cif family members on eukaryotic cell cycle progression. Shown is the flow cytometry analysis of DNA contents of EGFP-positive 293T cells transfected with indicated EGFP-tagged constructs.