Abstract
Altered growth and development of the endocrine pancreas is a frequent cause of the hyperglycemia associated with diabetes. Here we show that microRNA-375 (miR-375), which is highly expressed in pancreatic islets, is required for normal glucose homeostasis. Mice lacking miR-375 (375KO) are hyperglycemic, exhibit increased total pancreatic α-cell numbers, fasting and fed plasma glucagon levels, and increased gluconeogenesis and hepatic glucose output. Furthermore, pancreatic β-cell mass is decreased in 375KO mice as a result of impaired proliferation. In contrast, pancreatic islets of obese mice (ob/ob), a model of increased β-cell mass, exhibit increased expression of miR-375. Genetic deletion of miR-375 from these animals (375/ob) profoundly diminished the proliferative capacity of the endocrine pancreas and resulted in a severely diabetic state. Bioinformatic analysis of transcript data from 375KO islets revealed that miR-375 regulates a cluster of genes controlling cellular growth and proliferation. These data provide evidence that miR-375 is essential for normal glucose homeostasis, α- and β-cell turnover, and adaptive β-cell expansion in response to increasing insulin demand in insulin resistance.
Keywords: diabetes, glucagon, microRNA, islet, proliferation
The maintenance of β-cell mass during development and throughout life is a highly regulated process responsible for normal glucose homeostasis. Defects in the development of pancreatic islets lead to changes in islet composition, and they often result in the hyperglycemia that characterizes the diabetic state (1, 2). The dynamic adaptation of β-cell mass in adult life is influenced by various metabolic stresses, which control the balance between proliferation and apoptosis. These processes, known to be regulated at the transcriptional level, contribute to the development and maintenance of many tissues, including the pancreatic islet (3, 4). Recent studies have shown that micro RNAs (miRNAs), which regulate gene expression at a posttranscriptional level, are powerful regulators of growth, differentiation, and organ function (5–7). For instance, mutant mice in which miRNAs are collectively silenced during endocrine pancreas development exhibit defects in all pancreatic lineages, including a dramatic reduction of insulin-producing β cells (8). It is estimated that ≈30% of all protein coding genes are miRNA targets. Combining target prediction with experimental analysis of miRNA expression and production of loss of function mutants is beginning to improve our understanding of the roles that miRNAs play in normal and disease states (7–12). We have previously reported that miR-375, the highest expressed miRNA in pancreatic islets of humans and mice, regulates insulin secretion in isolated pancreatic β cells (13). In this study we have investigated the effect of genetic ablation of miR-375 on pancreatic islet development and function and in the etiology of type 2 diabetes.
Results
Development of Hyperglycemia in miR-375-Null Mice.
To elucidate the role of miR-375 in the maintenance of glucose homeostasis and the development of the pancreatic islet in vivo, we generated miR-375-null mice (375KO) by targeted deletion and homologous recombination in embryonic stem (ES) cells. The miR-375 gene is uniquely located within an intergenic region on mouse chromosome 1, and the targeting construct was designed to eliminate the entire ≈64-bp miRNA precursor sequence (Fig. S1A). Heterozygous mice were crossed and the mutants were confirmed by Southern blot analysis (Fig. S1B). Offspring of these intercrosses revealed genotypes of expected Mendelian ratios. An analysis of miR-375 by in situ hybridization confirmed its expression in wild-type and its absence in 375KO pancreatic islets (Fig. S1C). Northern blotting also confirmed the loss of expression in other neuroendocrine tissues in which miR-375 is expressed at low levels (Fig. S1D). MiR-375 null animals are fertile and exhibit no obvious abnormalities or changes in body mass (Fig. S1E).
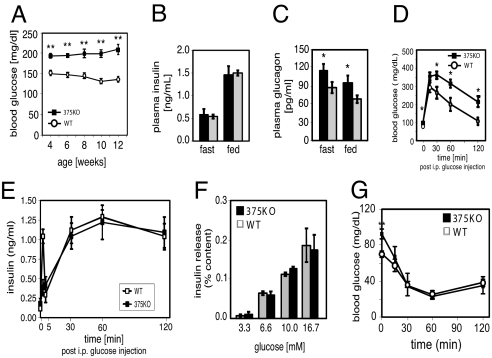
We investigated the metabolic consequences of miR-375 ablation by measuring fed and fasted glucose and islet hormone levels. At 4 weeks of age, male 375KO mice exhibited random hyperglycemia (Fig. 1A), and by 12 weeks they developed fasting hyperglycemia (89.7 mg/dL vs. 74.7 mg/dL, P < 0.001, 375KO vs. wild-type, respectively). Female 375KO mice developed random hyperglycemia by 8 weeks in the fed state. Despite the hyperglycemic state, plasma insulin levels remained unchanged in 375KO mice compared with wild-type littermates (Fig. 1B). In contrast, plasma glucagon concentrations were increased in both fasted and random-fed states (Fig. 1C). Mutant 375KO mice exhibit elevated glucose levels compared with wild-type controls after an i.p. glucose challenge (Fig. 1D). Under identical conditions, first-phase insulin release was diminished but plasma insulin levels were unchanged between 5 and 120 min after i.p. glucose administration (Fig. 1E). Glucose stimulation of isolated islets from 375KO and littermate control mice was similar over a range of concentrations (Fig. 1F). Furthermore, no significant differences in glucose clearance were measured during an insulin tolerance test, indicating the absence of peripheral insulin resistance (Fig. 1G).
Fig. 1.
miR-375-null mice develop diabetes. (A) Random-fed blood glucose levels in 375KO (filled squares) and wild-type littermate control (open circles) male mice. (B and C) Plasma insulin and glucagon levels in 10-week-old 375KO mice (black bars) and wild-type (gray bars) male mice. (D) Intraperitoneal glucose tolerance test administered to 10-week-old mice. (E) Plasma insulin levels during i.p. glucose tolerance test. (F) Insulin secretion of isolated islets in response to indicated glucose concentrations. (G) Insulin tolerance test of 375KO and wild-type littermates (n = 5).
We have previously shown that silencing of miR-375 increases glucose-stimulated insulin secretion in pancreatic β-cell lines and isolated primary β cells (13). To study the effect of chronic ablation of miR-375 on insulin secretion, we therefore measured exocytosis in single β cells by high-resolution capacitance measurements. Exocytosis was evoked by a train of depolarizations from −70 mV to 0 mV (Fig. S2). Responses were normalized to cell size. In wild-type cells, the exocytotic responses fell from an initial value of 6 fF/pF to 1.5 fF/pF at the end of the train. The total increase in capacitance during the train was 34 ± 5 fF/pF (n = 37). In β cells lacking miR-375, the exocytotic responses fell from an initial value of 7.5 fF/pF to 3.2 fF/pF and the total response evoked by the train amounted to 55 ± 6 fF/pF (P < 0.01 vs. wild-type; n = 46) (Fig. S2 A–C). An identical analysis was performed on isolated α cells; however, no differences were observed between mutant and wild-type animals (Fig. S2 D–F). These findings extend our earlier observations implicating miR-375 as a negative regulator of β-cell exocytosis (13). They also show that the hyperglycemia observed in 375KO mice is not due to a deficiency in insulin secretion.
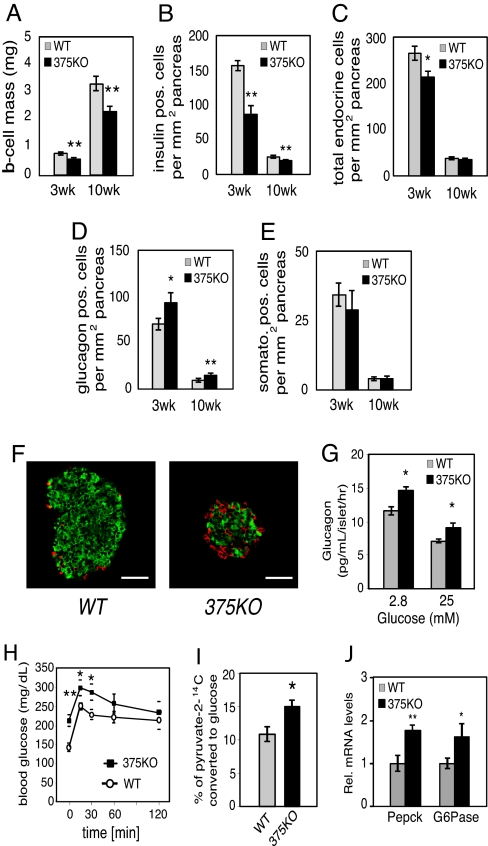
To further analyze the underlying cause for the metabolic derangements in 375KO mice, we investigated the endocrine pancreatic cell composition of mutant and control animals. Measurement of β-cell mass of 375KO pancreatic sections revealed a 38% and 31% decrease compared with wild-type controls at 3 and 10 weeks of age, respectively (Fig. 2A). Quantitative morphometric analysis of 375KO pancreatic sections from 3-week-old mice revealed that the change in mass was due to a comparable decrease in β-cell number (Fig. 2B) and resulted in a 20% decrease in total endocrine cells per pancreatic area compared with control mice (Fig. 2C). A similar decrease was observed in β-cell number at age 10 weeks in 375KO mice. In addition, these effects were accompanied by a 1.7-fold increase in α-cell number per pancreatic area compared with littermate controls (Fig. 2D). The number of δ cells was not changed in pancreata of 375KO mice compared with controls at either age (Fig. 2E). No changes in total pancreatic insulin or glucagon content, or pancreatic α- and β-cell numbers, were found at postnatal day (P)14. The results observed in 3-week-old animals are the earliest detectable changes in phenotype (Fig. 2 A–D). The morphological analysis also revealed disrupted islet architecture with increased presence of α cells within the islet core and in the periphery (Fig. 2F).
Fig. 2.
Decreased β-cell mass in 375KO pancreatic islets. (A) β-Cell mass in wild-type (gray bars) and 375KO (black bars) mice is quantified and reported as mean ± SE. (B–E) Quantification of endocrine cell number per total pancreatic area, β-cell number (B), total endocrine cell number per total pancreatic area (insulin, glucagon, and somatostatin-positive cells) (C), α-cell number (D), and δ-cell number (E) in 375KO (black bar) and wild-type (gray bar) male mice. (F) Representative sections of pancreas from 10-week-old 375KO and wild-type male mice visualized by immunofluorescence after staining with anti-insulin (green) and anti-glucagon (red) antibodies. (Bar, 50 μm.) (G) Glucagon secretion measured from islets isolated from 10-week-old male 375KO (black bars) and wild-type (gray bars) mice cultured overnight and incubated in fresh medium containing the indicated glucose concentrations. (H) Intraperitoneal pyruvate tolerance test was performed on random-fed 6-week-old male mice by administering a dose of sodium pyruvate (in saline) at 2 g/kg body weight. (I) [2-14C]Pyruvate was administered by i.p. injection into random-fed 6-week-old 375KO and wild-type (WT) mice and blood was drawn after 30 min and deproteinized, and labeled glucose in supernatant was recovered and radioactivity was measured. (J) Quantification of PEPCK and G6Pase mRNA expression by real-time PCR in liver from random-fed, 10-week-old 375KO (375KO) and wild-type (WT) mice. n = 5–12 animals per genotype unless otherwise noted. Data are presented as means ± SE. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To investigate if elevated plasma glucagon levels could explain the hyperglycemia in 375KO mice, we evaluated glucagon secretion and downstream effects in the liver. In contrast to glucose-stimulated insulin secretion, glucagon secretion was increased in isolated pancreatic islets of 375KO mice at both low (2.8 mM) and high (25 mM) glucose concentrations compared with wild-type littermates (Fig. 2G). Furthermore, pancreatic glucagon content was increased ≈3-fold compared with wild-type littermates (375KO vs. WT: 1.25 ± 0.28 vs. 0.41 ± 0.09 ng/mg tissue, P ≤ 0.01, n = 5). Hepatic glucose production was analyzed by measuring blood glucose levels after an i.p. injection of pyruvate in random-fed mice. Significantly higher plasma glucose levels at 15 and 30 min after injection indicated that 375KO mice have an increased ability to convert pyruvate to glucose compared with wild-type littermates (Fig. 2H). In addition, 375KO mice displayed a 25% increase in the rate conversion of [2-14C]pyruvate into blood glucose after i.p. injection, thereby providing further evidence that hepatic glucose production was increased in these mice (Fig. 2I). Similar results were obtained in fasted animals, demonstrating increased de novo synthesis of glucose by the liver in both fasted and fed conditions. Moreover, under random-fed conditions, real-time PCR analysis revealed a significant up-regulation of both phosphoenolpyruvate carboxykinase (PEPCK) and glucose 6-phosphatase (G6Pase) in the livers of 375KO compared with control mice, demonstrating that the hyperglucagonemia contributes to the elevated gluconeogenesis (Fig. 2J). Plasma and tissue levels of other neuroendocrine organs such as pituitary (growth hormone, cocaine- and amphetamine-regulated transcript), adrenal (noradrenaline, adrenaline, dopamine, corticosteroids) and intestinal (glucagon-like peptide 1, vasoactive intestinal peptide, secretin) peptides were similar in 375KO mice and littermate controls. In addition, challenging the mice with insulin after fasting and measuring ACTH and corticosterone to test the hypothalamic–pituitary–adrenal axis revealed no abnormality between mutant and wild-type animals, indicating that loss of miR-375 expression in the pituitary and adrenal does not contribute to the phenotype of the mutant mice. Taken together, these results show that the hyperglycemia measured in 375KO mice is primarily caused by hyperglucagonemia resulting from an increase in pancreatic α-cell mass.
Expression of miR-375 Is Required for Pancreatic β-Cell Compensation in Obesity.
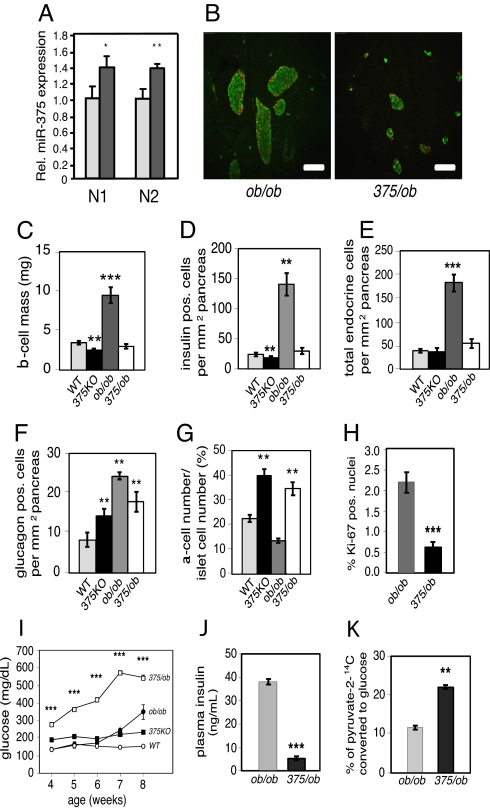
To further address the role of miR-375 in the maintenance of β-cell mass, we measured miR-375 expression in pancreatic islets isolated from ob/ob mice, a model for increased islet mass that is induced by severe insulin resistance (14). miR-375 expression was increased 30% in ob/ob islets compared with wild-type controls (Fig. 3A). We next generated mice deficient in both miR-375 and leptin (375/ob) to determine whether the increase in β-cell mass observed in ob/ob animals is dependent on miR-375 expression. Insulin and glucagon immunostaining from 10-week old 375/ob mice revealed an absence of islet hypertrophy compared with littermate control ob/ob mice (Fig. 3B). Pancreatic β-cell mass was decreased 71% and a similar reduction was measured in total β-cell number and total endocrine cell number per pancreatic area in 375/ob animals compared with ob/ob littermates (Fig. 3 C–E). The relative number of pancreatic α cells per area of pancreas was unchanged in 375/ob compared with 375KO animals (Fig. 3F). Consistent with 375KO mice, an increase in α-cell mass is reflected in an increase in the α- to β-cell ratio compared with both wild-type and ob/ob littermates (Fig. 3G). In addition, the decrease in β-cell number in 375/ob mice was accompanied by a decrease in β cells with Ki-67-positive nuclei (Fig. 3H). No changes were observed in ob/ob mice in which only 1 miR-375 allele was deleted. Failure of the islet mass to compensate for the insulin resistance induced by the obesity brought about a dramatic increase in blood glucose levels starting at age 4 weeks (Fig. 3I). Consistent with decreased β-cell mass, plasma insulin levels were decreased 85% in 375/ob animals compared with ob/ob mice (Fig. 3J) and plasma glucagon levels were unchanged (129.3 ± 7.5 pg/mL vs. 120.3 ± 10.1 pg/mL, 375/ob vs. ob/ob, respectively, n = 5–8). Furthermore, hepatic glucose production in 375/ob mice was elevated 1.8-fold compared with ob/ob mice (Fig. 3K). These results, in addition to the 40% decrease in body mass and measured polydipsia and polyuria, demonstrate severe insulin-deficient diabetes in 375/ob mice compared with ob/ob animals.
Fig. 3.
Impaired β-cell proliferation in miR-375/ob double-knockout mice. (A) Relative miR-375 expression in lepob/lepob (ob/ob) mice and wild-type (WT) controls measured by real-time PCR and normalized to U6 (N1) or miR-107 (N2) expression levels. (B) Representative 8-μm sections of pancreas from 10-week-old 375/ob (miR-375−/− leptin−/−) and ob/ob mice visualized by immunofluorescence after staining with insulin (green) and glucagon (red). (Bar, 50 μm.) (C) β-Cell mass in 10-week-old WT (gray bar), 375KO (black bar), ob/ob (dark gray bar), and 375/ob (open bar) mice is quantified and reported as mean ± SE. (D–F) Quantification of β-cell number (insulin-positive cells), total endocrine cell number (insulin, glucagon, and somatostatin-positive cells) and α-cell number (glucagon-positive cells) per total pancreatic area in wild-type (gray bars), 375KO (black bars), ob/ob (dark gray bars), and 375/ob (open bars) 10-week-old mice. (G) Ratio of α-cell number to islet cell number in wild-type (gray bar), 375KO (black bar), ob/ob (dark gray bar), and 375/ob (open bar) 10-week-old mice. (H) Quantification of percentage of Ki-67 insulin-positive nuclei within insulin-positive cells of 10-week-old 375/ob (black bar) and ob/ob (gray bar) mice. n > 30 for each genotype. (I) Random-fed blood glucose levels in 375/ob (open squares), ob/ob (filled circles), 375KO (filled squares), and wild-type littermate control (WT) (open circles) mice. (J) Plasma insulin levels in random-fed, 10-week-old 375/ob (black bar) and ob/ob (gray bar) mice. (K) Hepatic glucose production measured after sodium [2-14C]pyruvate was administered by i.p. injection into random-fed, 10-week-old 375/ob (black bar) and ob/ob (gray bar) mice. Data are presented as means ± SE. n = 4–6 animals per genotype unless otherwise noted. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
miR-375 Regulates Genes in Growth-Promoting Pathways.
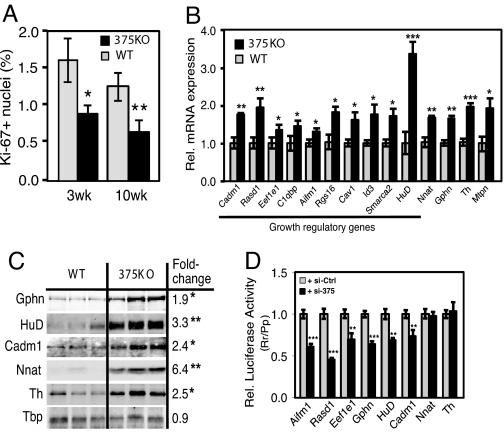
We next addressed whether the observed decrease in β-cell mass of 375KO mice could be reflective of changes in the rate of proliferation. Quantification of Ki-67-positive β cells, an index for cell proliferation, revealed a significant decrease in 375KO islets at 3 and 10 weeks of age (Fig. 4A). A similar result was obtained measuring BrdU incorporation in β cells of 375KO mice. To address the molecular basis for the decrease in pancreatic β-cell mass observed in the 375KO animals, we performed gene expression analysis by using Affymetrix microarrays comparing tissues from mutant mice to wild-type littermates. Four tissues expressing different levels of miR-375 were selected: pancreatic islets, pituitary, adrenal, and colon. Previous studies have established that miRNAs can negatively regulate the mRNA level of their direct targets (15), and that miRNA loss of function can result in the up-regulation of hundreds of genes (16). To determine the direct impact of loss of miR-375, we selected the most up-regulated 5% and the most down-regulated 5% of transcripts (see SI Methods). Each dataset thus contained 801 of the 16,301 Refseq transcripts measured by the array. We then determined the number of occurrences of the miR-375 recognition motif GAACAAA (corresponding to nucleotides 1–7 from the 5′ end of the miRNA) in the 3′UTRs of these transcripts. When measuring gene expression from pancreatic islets of 375KO mice compared with wild-type littermates, we counted 138 occurrences of the miR-375 motif in the dataset of up-regulated transcripts, and 49 occurrences in the dataset of down-regulated transcripts (Fig. S3A). Compared with random motifs with similar frequency across the 3′UTRs of all transcripts monitored by the array (represented in the graph by a blue box plot), the 138 up-regulated transcripts represent a 1.9-fold enrichment (P = 0.001), whereas the 49 down-regulated transcripts represent a 1.9-fold depletion (P = 0.002). These results demonstrate that genetic ablation of miR-375 in the pancreatic islet resulted in the up-regulation of direct targets of this miRNA. To further illustrate the impact of miR-375 on islet mRNA levels, we determined the distribution of expression changes of transcripts that do include a miR-375 motif in their 3′UTR and transcripts that do not. Transcripts that carry a miR-375 motif are up-regulated compared with transcripts that do not (P = 2.1 × 10−24 in the Wilcoxon rank-sum test), and the up-regulation is even stronger for transcripts containing evolutionarily selected miR-375 motifs (P = 0.005) (Fig. S3E). A similar analysis of gene expression in the pituitary of 375KO mice compared with wild-type littermates revealed a significant number of up-regulated motif-containing transcripts (Fig. S3C). By contrast, the genes up-regulated in the adrenal and colon data sets were not enriched for the miR-375 motif (Fig. S3 B and D). There are 2 possible explanations for this discrepancy: either the magnitude of the response from direct targets of miR-375 depends on the endogenous expression level of the miRNA, or miR-375 expression is limited to specific subpopulations of cells in the adrenal gland and colon. In situ hybridization using an miR-375-specific probe on pituitary tissue sections revealed miR-375 to be present in both the anterior and posterior pituitary, whereas its expression within the adrenal gland appears to be limited to the medulla and the zona glomerulosa of the cortex (Fig. S4). It is not known whether miR-375 is expressed in a specific cell type of the colon because probed tissue sections revealed no specific signal.
Fig. 4.
Regulation of gene expression and identification of growth target genes in 375KO islets. (A) Quantification of percentage of Ki-67-positive nuclei within insulin-positive cells of 375KO (black bars) and wild-type (gray bars) male mice. (B) Analysis of gene expression of putative miR-375 targets by real-time PCR in mutant and wild-type pancreatic islets. n = 5 animals per genotype. (C) Western blot analysis of protein lysates from pancreatic islets isolated from 375KO and wild-type (WT) male mice (100 islets per lane). Quantitative measurements made from densitometry are expressed as a ratio of mean values of 375KO to wild-type mice. (D) Increase in intracellular concentration of miR-375 decreases luciferase activity in HEK293 cells transfected with reporter constructs containing either full-length or partial 3′UTR sequence of putative miR-375 target genes (n = 6). Values relative to luciferase activity from cells transfected with a scrambled control are shown. Data are presented as means ± SE. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Several genes within the set of up-regulated transcripts of miR-375-null islets have been documented to negatively regulate cellular growth and were thus evaluated for direct regulation by miR-375. Selection of transcripts that contained a miR-375 recognition motif resulted in 381 putative direct targets of miR-375. Real-time PCR analysis confirmed 10 of these genes, including caveolin1 (Cav1), inhibitor of DNA binding 3 (Id3), Smarca2, Ras-dexamethasone-induced-1 (Rasd1), regulator of G protein signaling 16 (Rgs16), eukaryotic elongation factor 1 epsilon 1 (Eef1e1), apoptosis-inducing factor, mitochondrion-associated 1 (Aifm1), cell adhesion molecule 1 (Cadm1), HuD antigen (HuD), and complement component 1 q subcomponent binding protein (C1qbp) were up-regulated in 375KO islets (Fig. 4B). Increased expression of 3 additional genes, including cell adhesion molecule 1 (Cadm1), gephyrin (Gphn), and myotrophin (Mtpn), a previously validated target of miR-375 (13), was confirmed in 375KO islets by real-time PCR and Western blotting (Fig. 4 B and C). Furthermore, measurement of luciferase activity from HEK293 cells transfected with plasmid constructs containing a portion of or the entire 3′UTR of Aifm1, Rasd1, Eef1e1, Gphn, HuD, and Cadm1 showed reduced expression of all these constructs in the presence of miR-375 (Fig. 4D). These results suggest that Cav1, Id3, Smarca2, Aifm1, Rasd1, Rgs16, Eef1e1, C1qbp, HuD, and Cadm1, all of which have been shown to participate in signaling mechanisms that negatively regulate cellular growth and proliferation, are direct targets of miR-375. Published studies have shown that these genes play a role in the p53-dependent pathway (17–19), MAP kinase signaling (20), inducing apoptosis (21–23), and inhibiting normal developmental growth processes (24, 25) or the proliferation of tumors in mice (26, 27). Using real-time PCR analysis, we found that the expression levels of these genes in pancreatic islets either exceed or are comparable to the levels in tissues where a functional role has previously been determined (Fig. S5). We also confirmed changes in mRNA expression of several up-regulated genes that do not contain the miR-375 motif, including tyrosine hydroxylase (Th) and neuronatin (Nnat) (Fig. 4B). Although the exact role of these genes in the pancreatic β cell is not known, it was shown that increased expression of neuronatin is associated with hyperglycemia-induced apoptosis (28, 29). Both genes appear to be indirectly regulated by miR-375, because reporter assays with vectors that harbor their 3′UTRs downstream of the luciferase gene did not result in decreased activity when co-expressed with miR-375 (Fig. 4D). Together, these results provide evidence that many direct, as well as indirect, targets of miR-375 contribute to the regulation of the β-cell composition of islets.
Discussion
Our results illustrate an essential role for miR-375 in the establishment of normal pancreatic endocrine cell mass in the postnatal period and the maintenance of glucose homeostasis. The primary consequence resulting from the loss of miR-375 is chronic hyperglycemia caused by a pancreatic α-cell defect, as evidenced by increased α-cell mass, increased glucagon release from isolated islets, elevated fasted and fed plasma glucagon levels, and the increase in downstream effects of glucagon, such as expression of genes regulating gluconeogenesis and hepatic glucose production. Of note, 375KO mice in the fed state exhibit plasma glucagon levels that are comparable to fasted levels in wild-type mice, further emphasizing the chronic glucagon stimulus in these animals. The hyperglucagonemia in 375KO mice compared with control littermates is most likely due to the increase in α-cell numbers and a defect in glucose sensing because exocytosis measurements in isolated α cells in response to direct depolarization was similar in wild-type and mutant mice. The hyperglycemic phenotype of 375KO animals is unlikely due to the moderate (25%) decrease in β-cell mass because this reduction is usually insufficient to cause insulin deficiency and diabetes (30), and amounts of insulin secretion by isolated pancreatic islets from mutant and wild-type mice in response to various concentrations of glucose were similar. Furthermore, insulin levels in the fasted state and during a glucose challenge in 375KO and wild-type littermates were similar, despite a reduced β-cell number in 375KO mice, suggesting that insulin secretion per β cell is enhanced in 375KO mice and that reduction of β-cell mass and increased secretion balance each other in mutant mice.
The mechanism by which loss of miR-375 function leads to a reduced β-cell mass is most likely mediated by the cluster of negative growth regulators that are directly regulated by miR-375 and are markedly up-regulated in 375KO animals. The fact that the phenotype is more profound in mice with metabolic stress might indicate that miR-375 targets play a crucial role in β-cell compensation when metabolic demand is increased. The mechanism by which the α-cell number in 375KO pancreata is increased is currently unknown. Two models can be proposed: miR-375 regulates specific target genes in α cells that are responsible for increased α-cell mass. Alternatively, the increase in α-cell number could be the result of a compensatory response to altered β-cell mass and function or to the chronic hyperglucagonemia, which in some models is associated with α-cell hyperplasia (31, 32).
Mice bearing a conditional deletion of dicer, an enzyme required for miRNA processing, during pancreas development exhibit defects in all pancreatic cell lineages, abnormal islet architecture, and a profound reduction in pancreatic β cells (8). Mutant 375KO mice only partially mimic this phenotype, suggesting that miR-375 alone is not responsible for the marked developmental defect in β-cell growth and differentiation and that other miRNAs, which are expressed in endocrine pancreatic precursor cells, must be responsible for the observed phenotype of the Pdx-Cre/dicer mice.
Last, it is interesting that miR-375 plays a significant role in the hypertrophic growth response of pancreatic islets to metabolic stress. Expression levels of miR-375 are aberrant in obese mice, indicating that they contribute to increased β-cell mass in insulin resistance. Ablation of miR-375 expression in obese mice leads to a profound loss of β cells, metabolic decompensation, and premature death. Under these conditions, α-cell mass is not affected, suggesting that miR-375 has a less prominent role in α cells, which are not under particular metabolic or cellular stress in hyperglycemic/insulin resistant conditions. Increasing evidence implicates miRNAs as an essential component mediating responses to cellular stress. For instance, tissue-enriched miRNAs in the heart, such as miR-1, miR-208, and miR-133, have been shown to regulate the hypertrophic proliferative activity in response to a variety of stresses, and miR-126 affects survival after induction of a myocardial infarction (7, 12, 33). These observations from miRNA-knockout mice highlight the importance of small RNAs in cellular development, maintenance, and survival and reveal potential therapeutic targets for the treatment of disease.
Materials and Methods
Generation of 375KO and 375/ob Mice.
The murine miR-375 gene was deleted in Sv129 ES cells by homologous recombination by using a targeting vector in which the entire pre-miRNA was deleted and replaced by a dsRed cDNA and Neo selection cassette (Fig. S1A). Targeted clones were identified by BstEII digests of genomic DNA and Southern blotting using the indicated 3′ probe. Approximately 10% of clones carried the targeted allele and 2 clones were used to generate chimeric animals that passed the mutant allele to offspring (Fig. S1B). Double miR-375−/−, Lep−/− (375/ob) mice were generated by crossing double heterozygous mice and identified by PCR. Mice were housed in pathogen-free facilities in a 12-hr light/dark cycle and were backcrossed for 6 generations with C57BL/6 mice before characterization of animals. The dsRed transgene was not expressed. Unless stated, male animals were analyzed at 10 weeks of age.
Analysis of Metabolic Parameters.
Blood glucose, insulin, glucagon, free fatty acids, and triglycerides in plasma were measured as described (16, 34). Vasoactive intestinal polypeptide (VIP), cocaine- and amphetamine-regulated transcript (CART), and secretin were measured by RIA (Phoenix Pharmaceuticals). The following hormones were measured by ELISA: glucagon-like peptide 1 (Linco), cortisol (US Biological), and growth hormone (Diagnostic Systems). Catecholamines were measured from plasma and tissues by HPLC. Individual animals were placed in metabolic cages to measure water consumption and urinary output (Columbus Instruments).
Glucose, Insulin, and Pyruvate Tolerance Tests; in Vivo Gluconeogenesis, and Hypothalamic–Pituitary–Adrenal Axis Stimulation Studies.
Glucose tolerance tests were performed after mice were fasted overnight (16 hr) and injected i.p. with glucose (in saline) at 2 g/kg of body weight. Insulin tolerance tests were performed by injecting insulin i.p. (0.75 unit/kg of body weight), and measuring blood glucose before (time = 0) and 15, 30, and 60 min after injection. Pyruvate tolerance tests were also performed on mice in a random-fed state or after an overnight fast (16 hr) and injected i.p. with pyruvate (in saline) at 2 g/kg of body weight. In vivo gluconeogenesis studies were performed as previously described (35). Plasma corticosterone and ACTH were measured by RIA (Peninsula Laboratories and MP Biomedical, respectively).
Computational Analysis.
The expression analysis of total RNA extracted from tissues of 10-week-old animals by using TRIzol reagents (Invitrogen) was performed by using Affymetrix mouse genome 430 2.0 arrays. Analysis of total RNA extracted from MIN6 cells infected with recombinant adenovirus expressing miR-375 as described (13) was performed by using the Affymetrix mouse genome 430A array. Details on generation and analyses of data are found in SI Methods.
Northern Blotting, Quantitative PCR, Immunoblotting, and Luciferase Activity Measurements.
Northern blotting, Western blotting, and luciferase assays were performed as previously described (13). Antibodies for Western blotting were obtained from several different sources: anti-gephyrin (Chemicon), anti-igsf4a/cadm (R & D Systems), anti-neuronatin (Abcam), anti-tyrosine hydroxylase (Abcam), and anti-HuD (gift of R. Darnell, The Rockefeller University, New York, NY). Primer sequences for real-time PCRs are available on request. miRNA quantitative PCR results were normalized to U6 levels, which were measured by using the ABI miRNA U6 assay kit (Applied Biosystems).
Immunohistochemistry, Islet Morphometry, and in Situ Hybridization.
Immunohistochemistry was performed on at least 5 8-μm sections (at least 160 μm apart) prepared from paraffin-embedded pancreata of 3- and 10-week-old animals. Tissue sections were mounted with Vectashield with DAPI (Vector Laboratories) and analyzed by using a Leica DM5500 microscope, and the cross-sectional areas of pancreata and β cells (insulin-positive cells) were determined by using MetaMorph (version 7) software. Relative cross-sectional area of β cells was determined by quantification of the cross-sectional area occupied by β cells divided by the cross-sectional area of total tissue. β-Cell mass per pancreas was determined by the product of the relative cross-sectional area of β cells per total tissue and the pancreatic mass. Measurements were calculated by analyzing pancreata from at least 3 animals for each age and genotype. Cell quantification was based on counting nuclei of insulin-, glucagon-, or somatostatin-positive cells and data are represented as total cell number per pancreatic area. Ki-67- and BrdU-positive cells were counted from 1,500–2,000 insulin-positive cells per animal. Antibodies for immunofluorescence were obtained from several sources: anti-insulin and anti-glucagon (Linco), anti-somatostatin (Dako), anti-BrdU (Sigma), and anti-Ki-67 (Novocastra). BrdU incorporation and in situ hybridization were performed as described previously (9). Specific locked nucleic acid probes (Exiqon) were labeled by using terminal transferase and DIG-ddUTP (Roche).
Supplementary Material
Acknowledgments.
We thank J. Krutzfeldt, M. Spranger, C. Wolfrum, F. von Meyenn, M. Gstaiger, A. von Eckhardstein, A. Schaefer, R. Yi, and V. Horsley for advice and technical assistance. This work was supported in part by the Juvenile Diabetes Research Foundation, the National Institutes of Health, and the Wellcome Trust. M.N.P. was supported by a Ruth L. Kirschstein Fellowship. J.H. is partially supported by Schweizerische Nationalfonds Grant 3100A0-114001 to M.Z. M.T. was supported by a Juvenile Diabetes Research Foundation Post-Doctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810550106/DCSupplemental.
References
- 1.Edlund H. Pancreatic organogenesis—Developmental mechanisms and implications for therapy. Nat Rev Genet. 2002;3:524–532. doi: 10.1038/nrg841. [DOI] [PubMed] [Google Scholar]
- 2.Gromada J, Franklin I, Wollheim CB. alpha-Cells of the endocrine pancreas: 35 years of research but the enigma remains. Endocr Rev. 2007;28:84–116. doi: 10.1210/er.2006-0007. [DOI] [PubMed] [Google Scholar]
- 3.Shih DQ, et al. Profound defects in pancreatic beta-cell function in mice with combined heterozygous mutations in Pdx-1, Hnf-1alpha, and Hnf-3beta. Proc Natl Acad Sci USA. 2002;99:3818–3823. doi: 10.1073/pnas.062605899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson JD, et al. Increased islet apoptosis in Pdx1+/− mice. J Clin Invest. 2003;111:1147–1160. doi: 10.1172/JCI16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 8.Lynn FC, et al. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 9.Yi R, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 10.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 12.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a MicroRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 13.Poy MN, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 14.Bock T, Pakkenberg B, Buschard K. Increased islet volume but unchanged islet number in ob/ob mice. Diabetes. 2003;52:1716–1722. doi: 10.2337/diabetes.52.7.1716. [DOI] [PubMed] [Google Scholar]
- 15.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 16.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 17.Buckbinder L, et al. The p53 tumor suppressor targets a novel regulator of G protein signaling. Proc Natl Acad Sci USA. 1997;94:7868–7872. doi: 10.1073/pnas.94.15.7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galbiati F, et al. Caveolin-1 expression negatively regulates cell cycle progression by inducing G0/G1 arrest via a p53/p21(WAF1/Cip1)-dependent mechanism. Mol Biol Cell. 2001;12:2229–2244. doi: 10.1091/mbc.12.8.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park BJ, et al. The haploinsufficient tumor suppressor p18 upregulates p53 via interactions with ATM/ATR. Cell. 2005;120:209–221. doi: 10.1016/j.cell.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 20.Cohen AW, et al. Caveolin-1 null mice develop cardiac hypertrophy with hyperactivation of p42/44 MAP kinase in cardiac fibroblasts. Am J Physiol. 2003;284:C457–C474. doi: 10.1152/ajpcell.00380.2002. [DOI] [PubMed] [Google Scholar]
- 21.Kee BL. Id3 induces growth arrest and caspase-2-dependent apoptosis in B lymphocyte progenitors. J Immunol. 2005;175:4518–4527. doi: 10.4049/jimmunol.175.7.4518. [DOI] [PubMed] [Google Scholar]
- 22.Cheung EC, et al. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. EMBO J. 2006;25:4061–4073. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamal A, Datta K. Upregulation of hyaluronan binding protein 1 (HABP1/p32/gC1qR) is associated with cisplatin induced apoptosis. Apoptosis. 2006;11:861–874. doi: 10.1007/s10495-006-5396-4. [DOI] [PubMed] [Google Scholar]
- 24.Reyes JC, et al. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akamatsu W, et al. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc Natl Acad Sci USA. 2005;102:4625–4630. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuramochi M, et al. TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat Genet. 2001;27:427–430. doi: 10.1038/86934. [DOI] [PubMed] [Google Scholar]
- 27.Vaidyanathan G, et al. The Ras-related protein AGS1/RASD1 suppresses cell growth. Oncogene. 2004;23:5858–5863. doi: 10.1038/sj.onc.1207774. [DOI] [PubMed] [Google Scholar]
- 28.Borelli MI, Rubio M, Garcia ME, Flores LE, Gagliardino JJ. Tyrosine hydroxylase activity in the endocrine pancreas: Changes induced by short-term dietary manipulation. BMC Endocr Disord. 2003;3:2. doi: 10.1186/1472-6823-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joe MK, et al. Crucial roles of neuronatin in insulin secretion and high glucose-induced apoptosis in pancreatic beta-cells. Cell Signal. 2008;20:907–915. doi: 10.1016/j.cellsig.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Bonner-Weir S, Trent DF, Weir GC. Partial pancreatectomy in the rat and subsequent defect in glucose-induced insulin release. J Clin Invest. 1983;71:1544–1553. doi: 10.1172/JCI110910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker JC, Andrews KM, Allen MR, Stock JL, McNeish JD. Glycemic control in mice with targeted disruption of the glucagon receptor gene. Biochem Biophys Res Commun. 2002;290:839–843. doi: 10.1006/bbrc.2001.6265. [DOI] [PubMed] [Google Scholar]
- 32.Chen M, et al. Increased glucose tolerance and reduced adiposity in the absence of fasting hypoglycemia in mice with liver-specific Gs alpha deficiency. J Clin Invest. 2005;115:3217–3227. doi: 10.1172/JCI24196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulkarni RN, et al. beta-cell-specific deletion of the Igf1 receptor leads to hyperinsulinemia and glucose intolerance but does not alter beta-cell mass. Nat Genet. 2002;31:111–115. doi: 10.1038/ng872. [DOI] [PubMed] [Google Scholar]
- 35.Leveille GA, Chakrabarty K. In vivo and in vitro studies of gluconeogenesis in meal-fed and nibbling rats. J Nutr. 1968;96:397–402. doi: 10.1093/jn/96.3.397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.